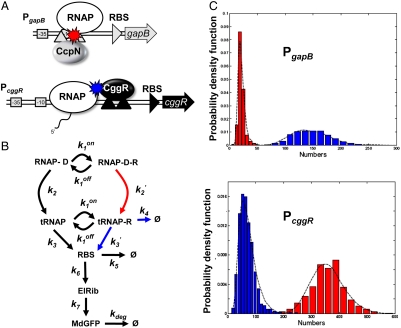
Fig. 4.
Model of gene regulation by CggR and CcpN. (A) Scheme describing the architecture of the B. subtilis PccgR and PgapB promoter region (boxed -10 and -35 RNAP recognition sequences) and tandem operator sites (black or gray upward triangles) for the CggR or CcpN repressors. Under glycolytic conditions, CcpN is thought to prevent promoter clearance by the RNA polymerase whereas under gluconeogenic conditions CggR acts as a roadblock to the transcribing polymerase when bound as a compact tetramer. (B) General model of prokaryotic gene expression and regulation applied to both repressors. RNAP-D is the RNAP-bound DNA, R the active repressor, tRNAP the elongating transcription complex, RBS the ribosome binding site on the transcribed mRNA, ElRib the elongating translation complex, and MdGFP the folded and matured green fluorescent protein. According to the above mechanistic models of regulation, besides changes in DNA affinity constants (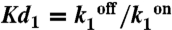 ), CcpN repression affects primarily k2, the rate at which the elongation complex is formed, whereas CggR would affect the transcription rate in the mRNA leader region (k3), thereby increasing the dissociation rate of the (paused) polymerase (k4). In the gfpmut3 reporter system used in this study, all steps past RBS production are identical for all promoter constructs and all conditions. The GFPmut3 variant has been shown to be fast-maturing (within a few minutes) and slow degrading (stable for several hours) in B. subtilis (3), therefore the degradation rate kdeg corresponds to slow dilution whereas the lifetime of the mRNA is much shorter (i.e., k5≫kdeg). (C) Results of the model compared to the experimental data for the stochastic expression of PgapBgfp and PcggRgfp transcriptional fusions under glucose (red) or malate (blue). Lines correspond to the continuous distributions obtained from the model parameters reported in Table S1. The histogram from the PgapBgfp promoter fusion data was not corrected for the BSB168 background contribution, as the deconvolution cannot be done reliably for experimentally reasonable dataset sizes; instead, a Gamma random variable having the same first two moments as the background contribution has been added to the model predictions.
), CcpN repression affects primarily k2, the rate at which the elongation complex is formed, whereas CggR would affect the transcription rate in the mRNA leader region (k3), thereby increasing the dissociation rate of the (paused) polymerase (k4). In the gfpmut3 reporter system used in this study, all steps past RBS production are identical for all promoter constructs and all conditions. The GFPmut3 variant has been shown to be fast-maturing (within a few minutes) and slow degrading (stable for several hours) in B. subtilis (3), therefore the degradation rate kdeg corresponds to slow dilution whereas the lifetime of the mRNA is much shorter (i.e., k5≫kdeg). (C) Results of the model compared to the experimental data for the stochastic expression of PgapBgfp and PcggRgfp transcriptional fusions under glucose (red) or malate (blue). Lines correspond to the continuous distributions obtained from the model parameters reported in Table S1. The histogram from the PgapBgfp promoter fusion data was not corrected for the BSB168 background contribution, as the deconvolution cannot be done reliably for experimentally reasonable dataset sizes; instead, a Gamma random variable having the same first two moments as the background contribution has been added to the model predictions.