Figure 1.
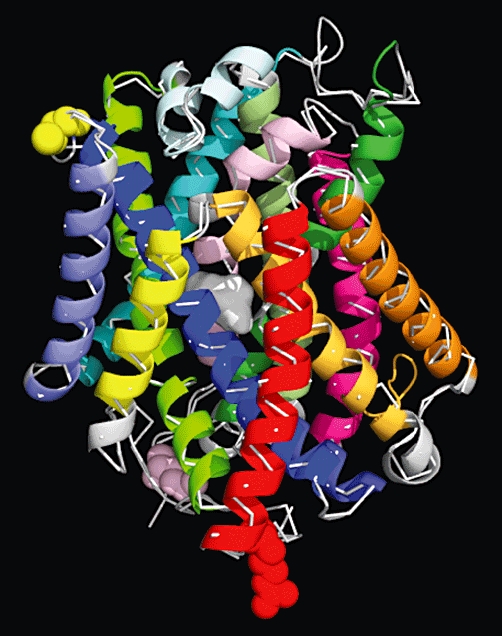
An xCT homology model was constructed by threading the hxCT sequence over the crystal structure of ApcT (RCSB: pdb3GIA) from M. jannaschii (GI:1591319) as reported by Goaux and co-workers (Shaffer et al., 2009) using FASTA (Pearson and Lipman, 1988) followed by multiple sequence alignments with ClustalW (Larkin et al., 2007). The structure of ApcT is shown as a white thread overlayed with the helical ribbons of the 12 TMDs of xCT: TMD 1A/B in light pink; TMD2 in dark pink; IL1 in dark blue; TMD3 in bright blue; TMD4 in light purple; TMD5 in teal, TMD6A/B in dark green; TMD7 in olive green; EL4A/B in silver; TMD8 in light green; TMD9 in yellow; TMD10 in gold; TMD11 in orange and TMD12 in red. Truncated intracellular N (light pink) and C (red) termini are shown with spheres. The cysteine-158 that participates in the disulphide bond with 4F2hc is highlighted with yellow spheres. The protein is shown in its inwardly facing Apo-form. A hypothetical substrate is depicted by the centrally located white surface as predicted by Schaffer et al., based upon the binding site of LeuT.
