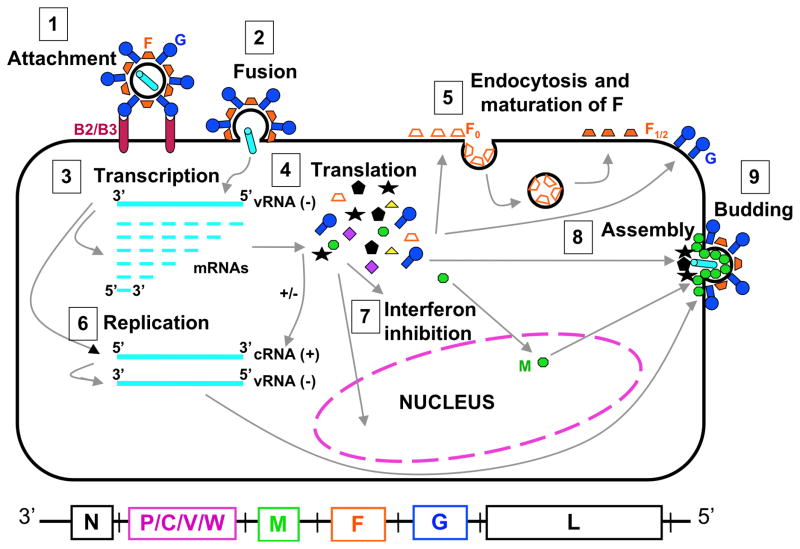
Fig. 2. Henipavirus replication cycle.
Depiction of henipavirus replication: After attachment to the B2/B3 receptor (1) and fusion (2), the virus enters the cell. The negative RNA genome (vRNA−) is a template for transcription of viral mRNAs following a 3–5′ attenuation gradient from N to L (3). mRNAs are translated into proteins (4) while the vRNA− is also a template for cRNA(+), which in turn is a template for vRNA(−) genomes during replication (6). New vRNA(−) genomes will be incorporated into new virions during viral assembly (8). Following translation (4), various viral proteins will function in interferon signaling pathways (7), and F0 will be endocytosed and matured (5). Assembly (8) and budding (9) are orchestrated primarily by the M protein, and N, P, C, M, F, and G, are incorporated into virions.