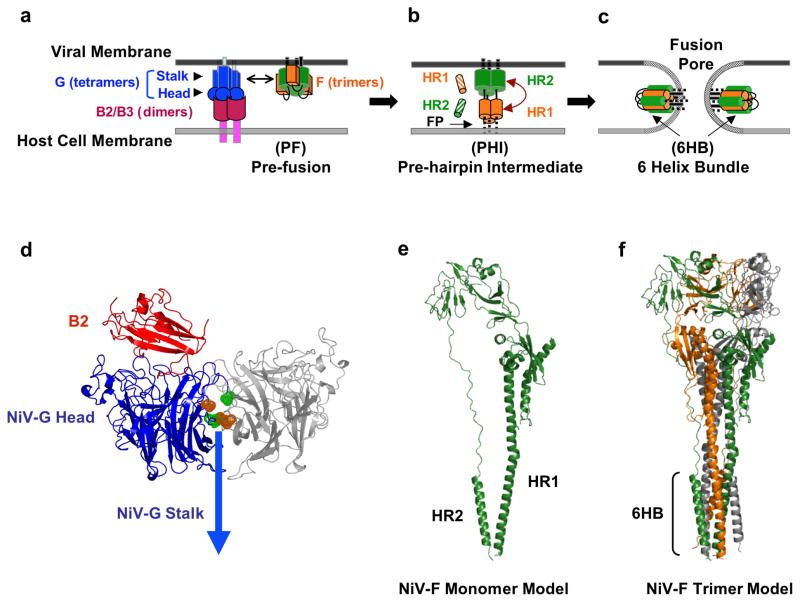
Fig. 3. Membrane Fusion and Viral Entry.
The attachment and membrane fusion steps necessary for viral entry (steps 1 & 2 from Fig. 2) are depicted here in greater detail in three major stages. (a) F is depicted in its pre-fusion, pre-hairpin intermediate, and post-fusion forms. EphrinB2 or ephrinB3 binding to NiV-G initiates a conformational cascade in F. (b) After F is triggered, it forms a pre-hairpin intermediate (PHI), in which a fusion peptide is harpooned into the host cell membrane. The PHI can be captured by peptides that mimic the NiV HR1 (orange striped cylinders) or HR2 regions (green striped cylinders) and bind the F HR2 or HR1 regions, respectively. (c) The HR1 and HR2 region in the PHI coalesce to form the six-helix bundle (6HB) conformation, bringing the viral and cell membranes together and facilitating viral-host membrane fusion and viral entry. At the figure bottom, the henipavirus genomic RNA is represented in its 3–5′ orientation. (d) Ribbon structure of the monomer of NiV-G (blue) head domain (pdb code 2VSM) and its interaction with its ephrinB2 receptor (red), drawn using PYMOL (www.pymol.org) and modeled by aligning the G/B2 monomer with each monomer of the hPIV3 Hemaglutinin-Neuraminidase dimer (pdb code 1V2I) similarly to (46). The second monomer is shown in gray. According to this model, the flexible region in the NiV-G ectodomain (green and orange) may interact with the same region in another monomer and may be involved in receptor-induced G mediated NiV-F triggering (46). (e) Representation of the structure of the NiV-F protein modeled using the HPIV3-F crystal structure (pdb code 1ztm) by the Phyre threding program, as performed in (78). (f) Representation of the trimer of NiV-F monomers from (e), also modeled using the HPIV3-F crystal structure as in (78).