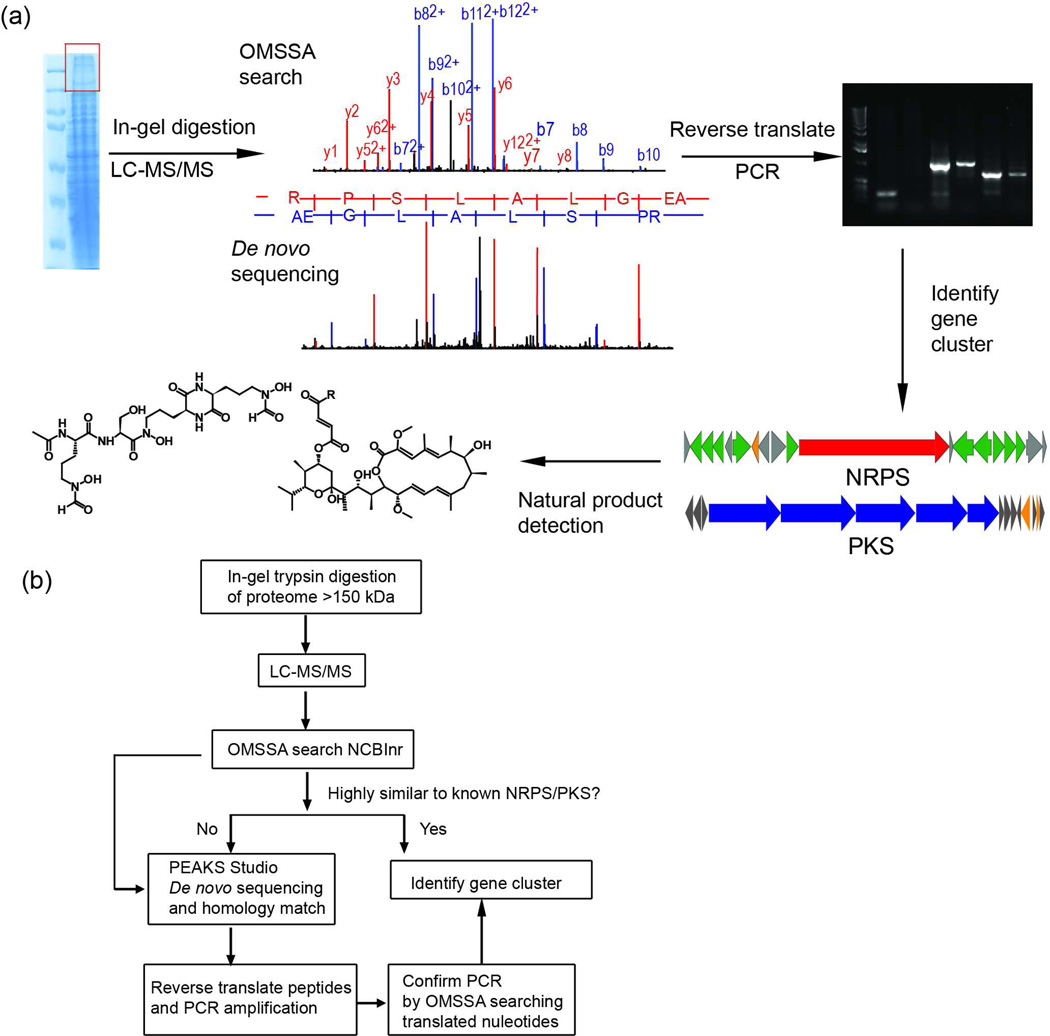
Figure 1.
(a) The work and logic flow for PrISM used in this study. High molecular weight proteins (>150 kDa) from a bacterial proteome are subjected to in-gel trypsin digestion and LC-MS/MS analysis. The data were analyzed by both database searching (top) and de novo peptide sequencing (bottom). Peptides identified as coming from NRPSs/PKSs were reverse translated into nucleotide sequences for PCR amplification of the target region. The PCR products guided the identification of biosynthetic gene clusters as well as discovery of the natural products. (b) Decision tree for LC-MS/MS data analysis. The LC-MS/MS data were first searched against NCBInr using OMSSA, and strains with NRPS/PKS identifications were divided into two categories based on their sequence similarity to known NRPS/PKS proteins, which was represented by the number of peptides identified and sequence coverage for each protein identification. Strains with low similarity to known NRPSs/PKSs were subjected to de novo peptide sequencing and homology-based searching by PEAKS Studio. Peptides identified by either search engines were used for PCR amplification. After confirming the specificity of the PCR products, they were used to guide the identification of biosynthetic gene clusters.