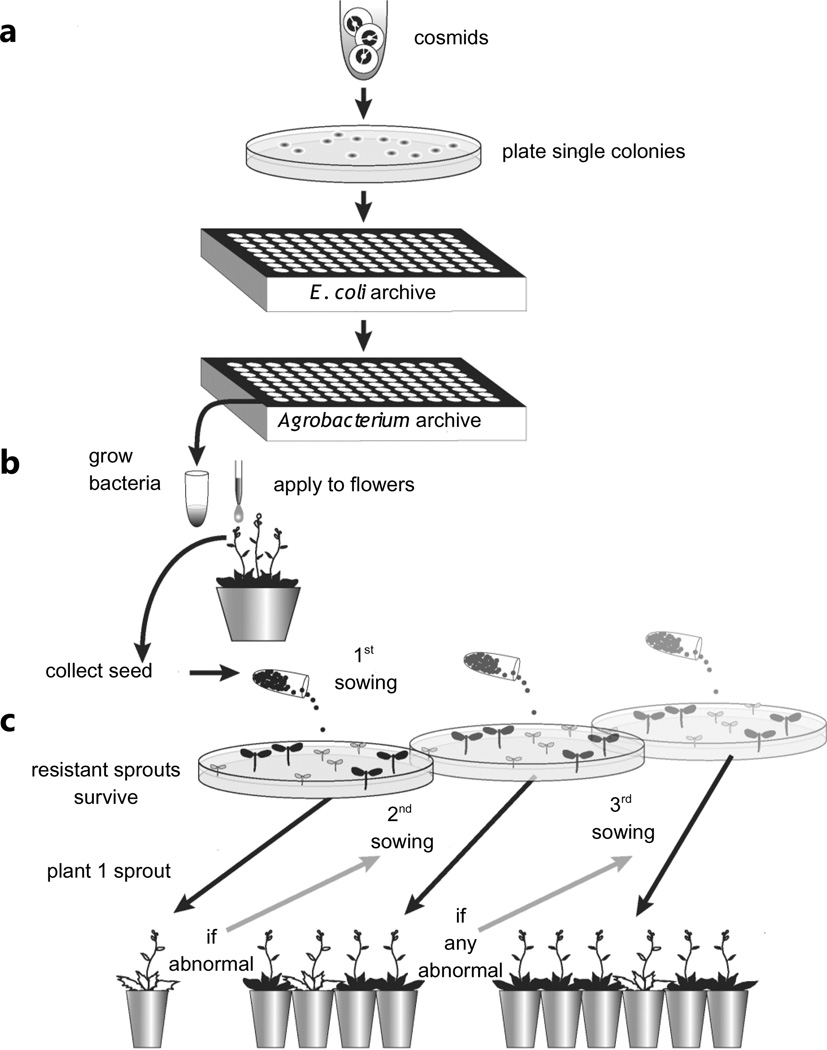
Fig. 2.
The clone-by-clone transgenomic strategy used to move Leavenworthia alabamica genomic DNA into Arabidopsis thaliana for phenotypic screening. (a) L. alabamica clones in E. coli are transferred 96-well format. Cosmids harboring L. alabamica inserts are then moved into Agrobacterium tumefaciens using high-throughput freeze thaw transformation. (b) A portion of each Agrobacterium stock in the 96-well plate is grown, then used to drip transform one pot of A. thaliana plants. Each T0 pot is then separately harvested for seed to identify primary transformant (T1) plants. (c) A portion of each T1 seed stock is then grown on kanamycin-containing plates. At least one kanamycin-resistant T1 plant is then transferred to a pot and allowed to grow to maturity. If the TI plant is abnormal, the transformed seed collection is sampled again to identify four additional independent T1 plants to see if the abnormal phenotype recurs. If the phenotype recurs, then a third seed sowing may be used to identify additional T1 plants.