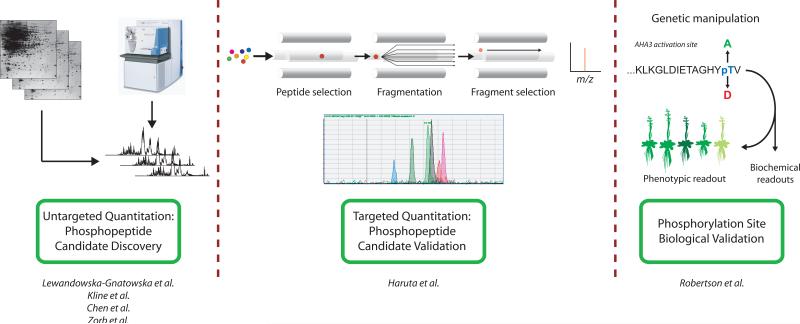
Figure 1.
Generalized quantitative phosphoproteomic workflow. Quantitative changes in phosphopeptide levels can be identified in biological samples using an untargeted mass spectrometry approach, such as done by Kline et al. [17], Zorb et al. [16], and Chen et al. [15]. Candidate phosphopeptides identified by the untargeted MS approach can then be targeted for further confirmation, analysis and quantitation using a targeted SRM-MS method, such as that shown by Haruta et al. [18]. Finally, site-specific genetic mutations and/or biochemical methods can be utilized to assess actual biological importance and impact of each phosphosite and/or phosphoprotein in the plant. [10]