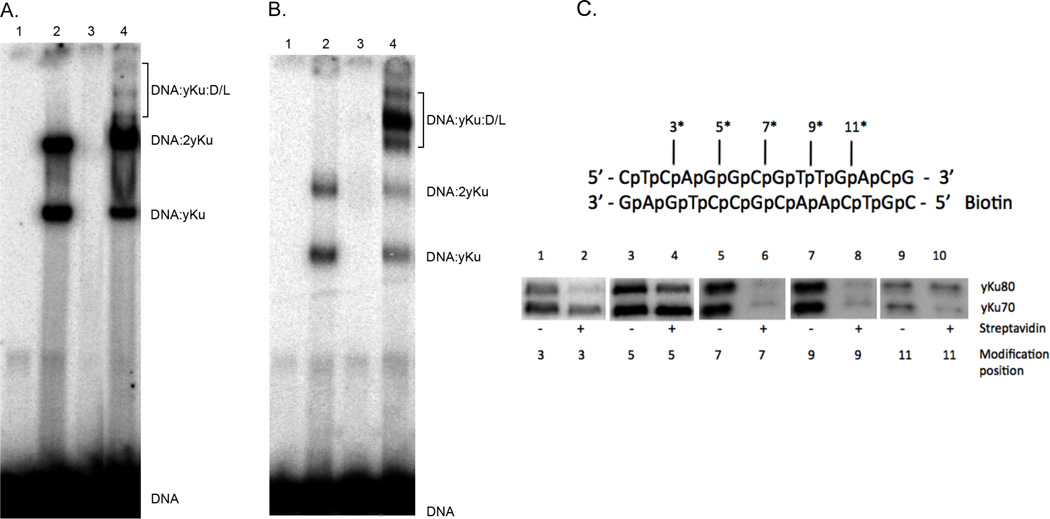
Figure 1. DNA-protein complex by yKu and Dnl4/Lif1 in EMSAs; Photocrosslinking of yKu to aryl azide-modified linear duplex DNA.
Radiolabeled 45 bp linear duplex DNA (25 nM) was incubated for 15 minutes at room temperature with; lane 1, no addition; lane 2, yKu (10 nM); lane 3, Dnl4/Lif1 (D/L, 20 nM) and lane 4, yKu (10 nM) and Dnl4/Lif1 (D/L, 20 nM). Incubation was continued for an additional 10 minutes at room temperature without (Panel A) or with 0.2% glutaraldehyde (Panel B). After separation by polyacrylamide (5%) gel electrophoresis, labeled DNA species in were visualized by phosphorImager analysis (Molecular Dynamics). The positions of the DNA substrate (DNA) and DNA-protein complexes formed with yKu (yKu) and Dnl4/Lif1 (D/L) are indicated. (C) Upper panel, schematic of the 14 bp DNA substrate used for photocrosslinking. The positions of phosphorothioate modifications within the top strand (asterisk) are indicated. Bottom panel, radiolabeled DNA substrates (7 nM) containing a single activated aryl azide modification within the top strand at the indicated distance from the end; lanes 1 and 2, 3 nucleotides; lanes 3 and 4, 5 nucleotides; lanes 5 and 6, 7 nucleotides; lanes 7 and 8, 9 nucleotides and lanes 9 and 10, 11 nucleotides. DNA (7 nM) and yKu (40 nM) were incubated with yKu (40 nM) in the presence or absence of streptavidin as indicated for 30 minutes at 25°C. Reactions were irradiated with 254 nM UV light for 2 minutes to trap DNA-protein complexes. After separation by 7.5% SDSPAGE electrophoresis, covalently-linked labeled DNA-protein complexes visualized by phosphorImager analysis.