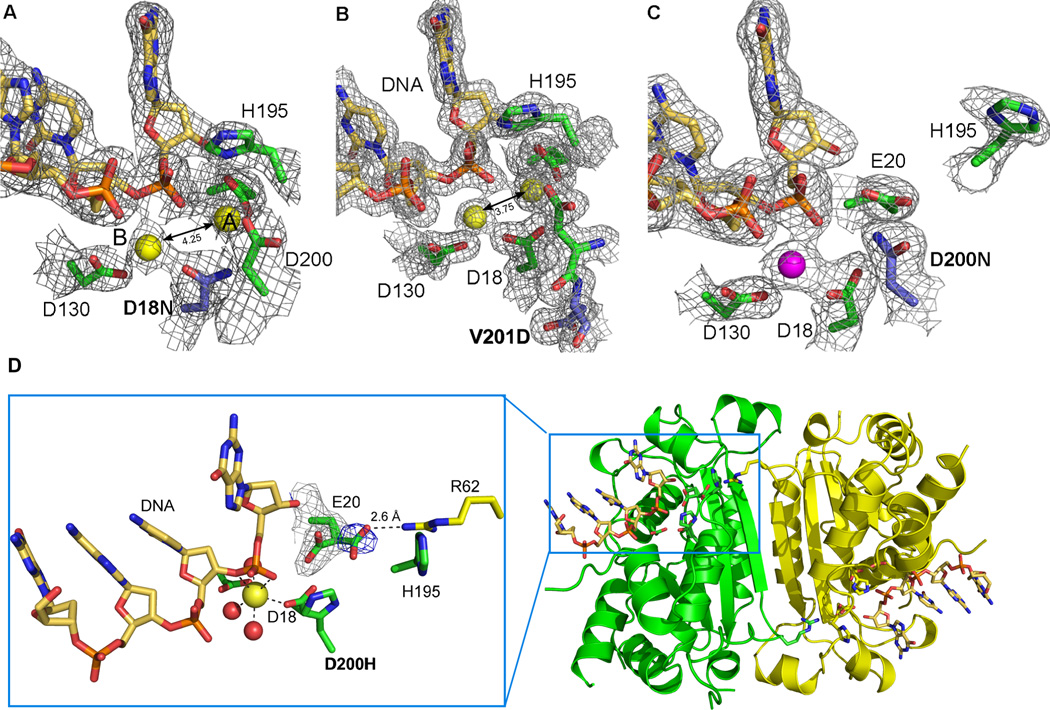
Figure 2. Active site morphology of TREX1 mutants.
The structures of the mutant TREX1 proteins fit into two distinct groups based on the number of metal ions coordinated and the position of residue H195 in the active sites. 2Fo-Fc electron density (1σ) for the D18N-DNA and V201D-DNA mutant TREX1 structures (A and B) reveal the catalytic H195 is pointing toward the active site and both divalent calcium ions (yellow spheres, canonical position A and B labeled) are present. However, the 4.25 Å and 3.75 Å interatomic distances for these metal ions is greater than the 3.5 Å distance needed for optimal catalytic activity. In contrast, the D200N-DNA and D200H-DNA structures (C and D) have the H195 oriented away from the active site and only a single divalent ion coordinated by the protein-DNA complex (Mg2+ shown as magenta sphere). The mutated residues in the AGS and FCL mutant active sites are highlighted in blue. (D) An alternate conformation of residue E20 is seen in the active site of the D200H mutant TREX1 structure (3.5 σ Fo-Fc density shown in blue) that allows an interaction with amino acid R62 from the opposing protomer in the dimer. This intersubunit interaction may play a role in substrate release and provides the first structural rationale for the dimeric nature of the TREX1 enzyme.