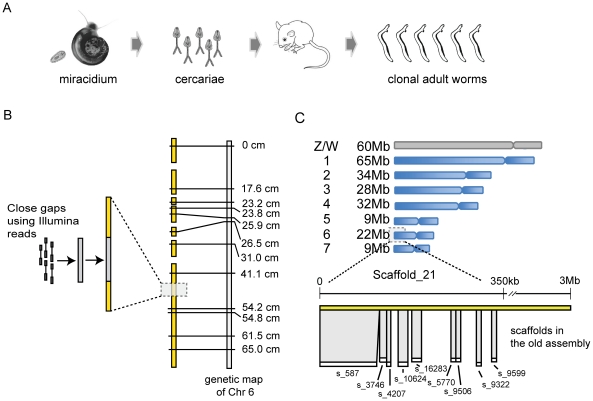
Figure 1. Improving the genome assembly of S. mansoni.
(A) Generation of clonal adult worms for Illumina sequencing. A single B. glabrata snail was infected with one miracidum only. The normal asexual reproduction stage of the sporocyst in the snail produces thousands of clonal cercariae that were used to infect mice. Clonal adult worms were recovered 7 weeks post-infection and processed for DNA extraction. (B) Closing gaps with IMAGE. Illumina data generated from the clonal adult worms were used to close gaps in the assembly using IMAGE [7] and, in conjunction with previous sequencing data, linkage markers and BAC ends, allowed the genome to be assembled into chromosomes. (C) Organisation of the S. mansoni genome into chromosomes. Top: The total length of the scaffolds that have evidence (either linkage markers or FISH-mapped BACs) assigning them to the 7 autosomal and W/Z chromosomes. Bottom: A schematic diagram showing the example of supercontig_21 (3 Mb), which was allocated to chromosome 6 using information from genetic mapping [17], and was able to link together 9 supercontigs from the old assembly into the first 350 kb.