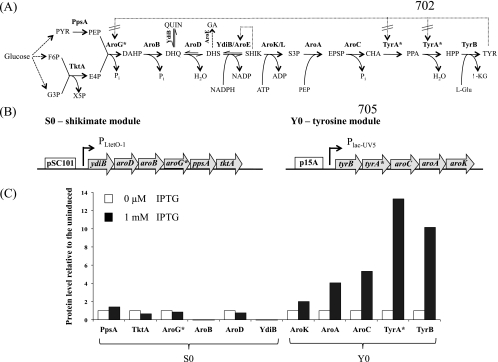
Fig 1.
(A) The biosynthetic pathway of l-tyrosine (TYR) in E. coli from glucose. X5P, xylulose 5-phosphate; PYR, pyruvate; EPSP, 5-enolpyruvoylshikimate 3-phosphate; CHA, chorismate; PPA, prephenate; HPP, 4-hydroxyphenlypyruvate; l-Glu, glutamic acid; and α-KG, α-ketoglutarate. The enzymes (in boldface) are as follows: PpsA, phosphoenolpyruvate synthase; TktA, transketolase A; AroG, DAHP synthase; AroB, DHQ synthase; AroD, DHQ dehydratase; YdiB, quinate/shikimate dehydrogenase; AroE, shikimate dehydrogenase; AroK/L, shikimate kinase I/II; AroA, EPSP synthase; AroC, chorismate synthase; TyrA, chorismate mutase/prephenate dehydrogenase; and TyrB, tyrosine aminotransferase. QUIN and gallic acid (GA) are side products. QUIN is formed by YdiB from DHQ (18), while GA is formed by AroE from DHS (18, 30). The dashed lines indicate where feedback inhibitions occur. Allosteric regulation of AroG and TyrA were removed in this study by employing their respective feedback-resistant mutants, AroG* (D146N) and TyrA* (M53I;A354V), respectively. (B) Structures of the initial modules, S0 and Y0, for production of shikimate and l-tyrosine, respectively. The open blocks indicate the origins of replication, the shaded arrows represent the genes, and the angled arrows indicate the promoters. Note that for each operon, the genes are placed in the reverse order relative to the reaction pathway. Using the shikimate module as an example, ydiB, which catalyzes the last step in the formation of shikimate, was placed next to the promoter, and so on. (C) SRM analysis of the protein production levels from S0 and Y0 in strain A when induced or uninduced with IPTG. The protein levels shown are ratios relative to the uninduced levels.