Abstract
A combined molecular and cultural method for the detection of the Mycobacterium tuberculosis complex (MTBC) and Mycobacterium avium subsp. paratuberculosis was developed and tested with artificially contaminated milk and dairy products. Results indicate that the method can be used for a reliable detection as a basis for first risk assessments.
TEXT
Tuberculosis in cattle, the most important known source of human food-borne tuberculosis infections, is predominantly associated with Mycobacterium bovis and Mycobacterium caprae. Many European countries have been recognized officially as free of bovine tuberculosis (OTF), but this status does not necessarily imply national herds being totally free of bovine tuberculosis (4, 5). At present, food matrices are not routinely checked for the presence of the Mycobacterium tuberculosis complex (MTBC). Food safety depends mainly upon veterinary inspections during slaughtering and the pasteurization technology in the milk sector. These measurements cover the majority of products sold on the food market. Nevertheless, they can fail as an adequate control of special products, like raw milk cheese. Mycobacterium avium subsp. paratuberculosis, which may be implicated in Crohn's disease (CD) in humans, is also associated mainly with cattle and small ruminants (Johne's disease). At the moment, a causative role of M. avium subsp. paratuberculosis in the etiology of CD is widely discussed (12), but for a well-founded risk assessment also concerning the food-borne transmission, it is necessary to collect data about the prevalence in different food matrices. The aim of our study was to test different cultural methods in combination with a real-time PCR assay for the detection of MTBC and M. avium subsp. paratuberculosis in milk and dairy products. The efficiency of the method was monitored with artificially contaminated samples.
The freely available Primer3 software (http://frodo.wi.mit.edu) was used for primer and probe design of the helicase gene of the MTBC. The theoretical specificity was checked with the BLAST database first. Then this primer and probe set was combined with a heterologous internal amplification control as described previously (10) and the detection system amplifying the F57 and the ISMav2 gene sequences of M. avium subsp. paratuberculosis described by Schönbrücher et al. (14) (Table 1). A temperature-time profile, with an initial denaturation step at 95°C for 10 min and 45 cycles with a denaturation step at 95°C for 15 s and an annealing step at 55°C for 60 s, was adapted for the Stratagene MX 3000P and MX 3005P thermocyclers (Agilent Technologies). The primers were used with a concentration of 300 nM and the probes with a concentration of 100 nM per reaction in combination with the Brilliant Multiplex MasterMix (Agilent Technologies). DNA extraction from isolates was done with thermal lysis (95°C for 10 min). For the DNA extraction from enrichment broths, the Maxwell purification system (Promega, Germany) was used after centrifugation at 10,000 × g for 5 min and lysis of the resulting pellet at 37°C for 30 min using a 10 mM Tris-EDTA buffer (pH 8) with a concentration of 3 mg/ml of lysozyme (Carl Roth, Germany). The specificity of the system was tested with 47 bacterial, yeast, and fungal strains (DSMZ, ATCC, and field strains, isolated and characterized at our laboratory). For the determination of robustness and sensitivity, standard curves using DNA serial dilutions were generated. The quantification for further tests was done with a most probable number (MPN) technique in accordance with the U.S. Food and Drug Administration Bacteriological Analytical Manual (1). The MPN was calculated from 56-day-enriched cultures of the bacillus calmette-guérin (BCG) strain (ATCC 27289) and an M. avium subsp. paratuberculosis strain (ATCC 19698), plated on Middlebrook agar plates (BD Diagnostics) for the BCG strain and Herold's egg yolk agar with mycobactin (BD Diagnostics) for the M. avium subsp. paratuberculosis strain and also investigated with real-time PCR. For further tests, the CFU per milliliter of enriched culture was calculated depending on the positive real-time PCR results. For the cultural detection, Kirchner broth and Dubos broth (BD Diagnostics) were tested with serial dilutions using different enrichment conditions and with artificially contaminated milk, yoghurt, sour cream, and curd cheese samples. Every 10 g of sample material was contaminated with three different contamination levels (104 CFU, 103 CFU, and 102 CFU). The test portions were incubated under aerobic conditions at 37°C for 7, 14, and 56 days. At every date, the enrichment broths were cultured onto Middlebrook agar plates (MTBC) or onto Herold's egg yolk agar with mycobactin (M. avium subsp. paratuberculosis). The agars were incubated for 7 to 56 days, depending on the presence of typical colonies. Real-time PCR directly from the enrichment broths was done before incubation and after 7, 14, and 56 days.
Table 1.
Primer and probe sequences for the detection of MTBC and M. avium subsp. paratuberculosis
| Name | Sequence (5′–3′) | Modificationa | Gene (GenBank accession no.) | Source or reference |
|---|---|---|---|---|
| Myco_tub_fw | GAA CCC GCT GAT GCA AGT | M. bovis helicase gene, species-specific sequence (U87961.1) | This study | |
| Myco_tub_re | GCG AAC CCT ACC TAC ATC GT | M. bovis helicase gene, species-specific sequence (U87961.1) | This study | |
| Myco_tub_S_ | CTG ACG GTG GTC ACC TTC TT | FAM-TAMRA | M. bovis helicase gene, species-specific sequence (U87961.1) | This study |
| Myco_para_F57_fw | TAC GAG CAC GCA GGC ATT C | M. avium subsp. paratuberculosis F57 (X70277) | 14 | |
| Myco_para_F57_re | CGG TCC AGT TCG CTG TCA T | M. avium subsp. paratuberculosis F57 (X70277) | 14 | |
| Myco_para_F57_S | CCT GAC CAC CCT TC | HEX-TAMRA | M. avium subsp. paratuberculosis F57 (X70277) | 14 |
| Myc_par_ISMav_fw | CGG CAA AAT CGA GCA GTT TC | M. avium subsp. paratuberculosis ISMav2 (AF286339) | 14 | |
| Myc_par_ISMav_re | TGA GCC GGT GTG ATC ATC TT | M. avium subsp. paratuberculosis ISMav2 (AF286339) | 14 | |
| Myc_pa_ISMav_S | CGC TGA GTT CCT TAG | Cy5-BHQ2 | M. avium subsp. paratuberculosis ISMav2 (AF286339) | 14 |
| pUC_19_fw | TGT GAA ATA CCG CAC AGA TG | pUC 19 (L09137) | 10 | |
| pUC_19_re | AGC TGG CGT AAT AGC GAA G | pUC 19 (L09137) | 10 | |
| pUC_19_S | GAG AAA ATA CCG CAT CAG GC | ROX-TAMRA | pUC 19 (L09137) | 10 |
FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine; HEX, 5′-hexachlorofluorescein; ROX, carboxy-X-rhodamine.
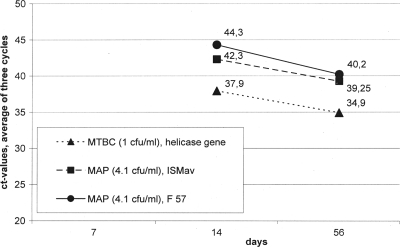
The real-time PCR systems showed 100% specificity for the panel of strains tested. No cross-reaction was found between the MTBC and the M. avium subsp. paratuberculosis detection systems. The sensitivity was assigned with 100 fg (about 20 genome equivalents) for the detection of MTBC and M. avium subsp. paratuberculosis. The robustness test showed a maximum difference of 1.2 cycles in different real-time PCR runs and on different thermocyclers. When Dubos broth and Kirchner broth were tested with pure cultures and artificially contaminated samples, both broths showed no differences in the detection limits for BCG and M. avium subsp. paratuberculosis. The detection limits using pure cultures for the enrichment broths in combination with the real-time PCR system are shown in Fig. 1. Using only cultural methods, the detection limit was 1,000 CFU/ml for the BCG strain and 410 CFU/ml for the M. avium subsp. paratuberculosis strain after 14 and 57 days of enrichment. The results using artificially contaminated samples are shown in Tables 2 and 3. The detection with only cultural methods failed for samples contaminated with less than 103 CFU/g sample material. But in every case the differences in the threshold cycle (CT) values of the real-time PCR results showed an increase of DNA during the incubation period (more than 2.4 cycles, double the value of the robustness test). Therefore, it can be assumed that there were viable and culturable bacteria in the artificially contaminated samples. Our investigations showed that only a molecular screening combined with the possibility of a cultural detection in a positive case can be used for a reliable detection of MTBC and M. avium subsp. paratuberculosis in milk and dairy products on the retail level. The risk of MTBC as a reemerging pathogen entering the food chain and causing human infection is clearly demonstrated by Harris et al. (7). Therefore, the need for reliable detection and risk-based investigations of different matrices is obvious in the framework of the official food control. Present reports about the development of molecular methods for the detection and differentiation of MTBC deal with clinical human and veterinary samples (3, 9, 11). They cannot be transmitted for the investigation of food, as low contamination levels per gram of sample material are to be expected in these matrices. In contrast to MTBC, many recent reports dealt with M. avium subsp. paratuberculosis in milk and dairy products (2, 6, 8, 15). Not all studies were carried out with combined cultural and molecular methods; some of them dealt only with PCR or real-time-PCR-positive samples (8). For a solid risk assessment concerning, e.g., heat-treated dairy products like infant formula, data about the viability of M. avium subsp. paratuberculosis are decisive. RNA-based molecular methods developed for a differentiation between viable and dead cells (2a) might provide important information about the prevalence in milk and dairy products, but cultural data on the retail level are also needed. Therefore, the combined cultural and molecular method presented in our study can be useful for a routine screening of milk and dairy products for MTBC and M. avium subsp. paratuberculosis on the retail level as a basis for a preliminary risk assessment.
Fig 1.
Detection limit for MTBC and M. avium subsp. paratuberculosis using enrichment broths in combination with real-time PCR.
Table 2.
Investigation of artificially MTBC-contaminated samples using an enrichment procedure in combination with real-time PCR
| Enrichment time (no. of days) |
CT values (avg of 3 PCR runs) for different contamination levels (CFU/g sample material)a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Milk (3.8% fat) |
Yoghurt |
Sour cream |
Curd cheese (40% fat) |
|||||||||
| 103 | 102 | 101 | 103 | 102 | 101 | 103 | 102 | 101 | 103 | 102 | 101 | |
| 0 | 36.0 | 39.5 | – | 40.2 | 44.3 | – | 42.4 | 43.5 | – | 40.4 | – | – |
| 7 | 28.9 | 32.7 | 39.9 | 29.7 | 39.3 | – | 32.0 | 36.3 | 40.2 | 31.1 | – | – |
| 14 | 27.3 | 30.6 | 36.7 | 29.3 | 38.5 | 43.2 | 31.5 | 36.0 | 40.0 | 30.7 | – | – |
| 56 | 22.4 | 28.4 | 34.3 | 26.2 | 34.2 | 40.0 | 28.2 | 34.2 | 36.7 | 27.3 | 40.2 | – |
–, negative value.
Table 3.
Investigation of artificially M. avium subsp. paratuberculosis-contaminated samples using an enrichment procedure in combination with real-time PCR
| Enrichment time (no. of days) |
CT values (avg of 3 PCR runs) for different contamination levels (CFU/g sample material)a |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Milk (3.8% fat) |
Yoghurt |
Sour cream |
Curd cheese (40% fat) |
|||||||||
| 103 | 102 | 101 | 103 | 102 | 101 | 103 | 102 | 101 | 103 | 102 | 101 | |
| 0 | 38.8/40.5 | 42.4/44.3 | –/– | 39.9/42.3 | –/– | –/– | 41.1/44.3 | –/– | –/– | –/– | –/– | –/– |
| 7 | 36.8/39.5 | 41.4/42.3 | –/– | 34.8/37.3 | –/– | –/– | 35.8/38.3 | 40.3/43.1 | –/– | 38.6/42.2 | –/– | –/– |
| 14 | 35.7/37.3 | 39.3/40.2 | –/– | 33.7/36.1 | –/– | –/– | 35.8/38.3 | 40.3/43.1 | –/– | 36.8/40.1 | –/– | –/– |
| 56 | 33.4/36.5 | 37.2/39.3 | 40.2/42.7 | 31.4/33.4 | 37.6/35.2 | 40.8/43.2. | 35.8/38.3 | 40.3/43.1 | 43.3./44.5 | 31.4/36.7 | 37.8/40.2 | –/– |
–, negative value. The first of each pair of values is from the F57 gene detection system; the second is from the ISMAv2 gene detection system.
Footnotes
Published ahead of print 4 November 2011
REFERENCES
- 1. Blodgett R. 2003. Most probable number from serial dilutions. In Bacteriological analytical manual, 8th ed, appendix 2. US Food and Drug Administration, Washington, DC. http://www.fda.gov [Google Scholar]
- 2. Botsaris G, et al. 2010. Rapid detection methods for viable Mycobacterium avium subspecies paratuberculosis in milk and cheese. Int. J. Food Microbiol. 141(Suppl. 1):87–90 [DOI] [PubMed] [Google Scholar]
- 2a. Dzieciol M, et al. 2010. A novel real-time PCR assay for specific detection and quantification of Mycobacterium avium subsp. paratuberculosis in milk with the inherent possibility of differentiation between viable and dead cells. BMC Res. Notes 3:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ereqat S, et al. 2010. Rapid differentiation of Mycobacterium tuberculosis and M. bovis by high-resolution melt curve analysis. J. Clin. Microbiol. 48:4269–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. European Food Safety Authority 2003. Opinion of the scientific panel on biological hazards on a request from the commission related on “Tuberculosis in bovine animals: risks for human health and control strategies.” EFSA J. 13:1–53 [Google Scholar]
- 5. European Food Safety Authority 2010. The community summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in the European Union in 2008. EFSA J. 8:1496 [Google Scholar]
- 6. Gill CO, Saucier L, Meadus WJ. 2011. Mycobacterium avium subsp. paratuberculosis in dairy products, meat, and drinking water. Food Prot. 74:480–499 [DOI] [PubMed] [Google Scholar]
- 7. Harris NB, et al. 2007. Recovery of Mycobacterium bovis from soft fresh cheese originating in Mexico. Appl. Environ. Microbiol. 73:1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hruska K, Slana I, Kralik P, Pavlik I. 2011. Mycobacterium avium subsp. paratuberculosis in powdered infant milk: F57 competitive real time PCR. Vet. Med. 56(5): 226–230 [Google Scholar]
- 9. Kumar P, et al. 2009. Visual format for detection of Mycobacterium tuberculosis and M. bovis in clinical samples using molecular beacons. J. Mol. Diagn. 11:430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Messelhäusser U, et al. 2007. Real-time-PCR for the detection of emetic Bacillus cereus in food. J. Verbr. Lebensm. 2:190–193 [Google Scholar]
- 11. Parra A, et al. 2008. Development of a molecular diagnostic test applied to experimental abattoir surveillance on bovine tuberculosis. Vet. Microbiol. 127:315–324 [DOI] [PubMed] [Google Scholar]
- 12. Rosenfeld G, Bressler B. 2010. Mycobacterium avium paratuberculosis and the etiology of Crohn's disease: a review of the controversy from the clinician's perspective. Can. J. Gastroenterol. 24:619–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reference deleted.
- 14. Schönenbrücher H, Abdulmawjood A, Failing K, Bülte M. 2008. New triplex real-time PCR assay for detection of Mycobacterium avium subsp. paratuberculosis in bovine feces. Appl. Environ. Microbiol. 74:2751–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shankar H, et al. 2010. Presence, characterization, and genotype profiles of Mycobacterium avium subspecies paratuberculosis from unpasteurized individual and pooled milk, commercial pasteurized milk, and milk products in India by culture, PCR, and PCR-REA methods. Int. J. Infect. Dis. 14:121–126 [DOI] [PubMed] [Google Scholar]