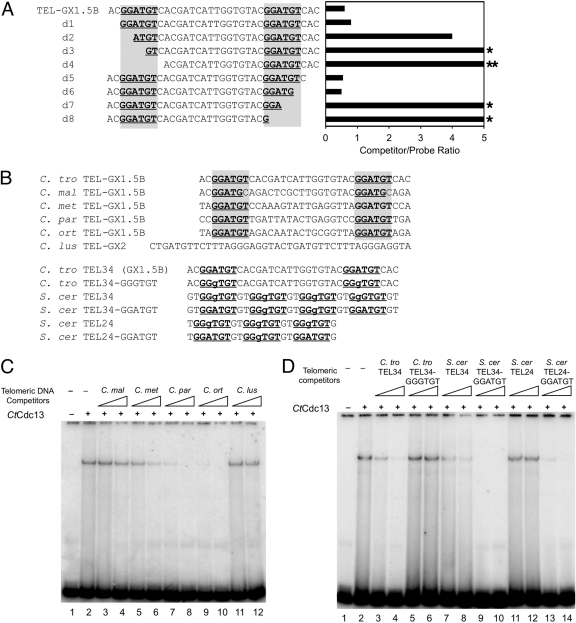
Fig 4.
The role of a 6-nucleotide element in CtCdc13-DNA interaction. (A) The listed oligonucleotides were used as competitors in EMSAs containing 10 nM CtCdc13 and 7.5 nM labeled TEL-GX2 DNA. The locations of the GGATGT sequence element required for high-affinity binding are highlighted. The amount of competitor oligonucleotide required to achieve a 50% reduction in complex formation was determined and plotted. Stars designate cases where a 4-fold excess competitor resulted in ∼30% and ∼0% reductions in complex formation, respectively. (B) The sequences of oligonucleotides used as competitors in EMSAs are displayed. The consensus GGATGT and the closely related GGgTGT elements are highlighted. (C) The indicated oligonucleotides (consisting of various Candida telomere repeats: C. maltosa [C. mal], C. metapsilosis [C. met], C. parapsilosis [C. par], C. orthopsilosis [C. ort], and C. lusitaniae [C. lus]) were used as competitors in EMSAs that contained 10 nM CtCdc13 and 7.5 nM labeled TEL-GX2 DNA. The competitors were added at levels 2.5- and 10-fold higher than that for the probe. (D) The indicated oligonucleotides (consisting of wild-type or mutated S. cerevisiae [S. cer] and C. tropicalis [C. tro] telomere repeats) were used as competitors in EMSAs that contained 10 nM CtCdc13 and 7.5 nM labeled TEL-GX2 DNA. The competitors were added at levels 2.5- and 10-fold higher than that for the probe.