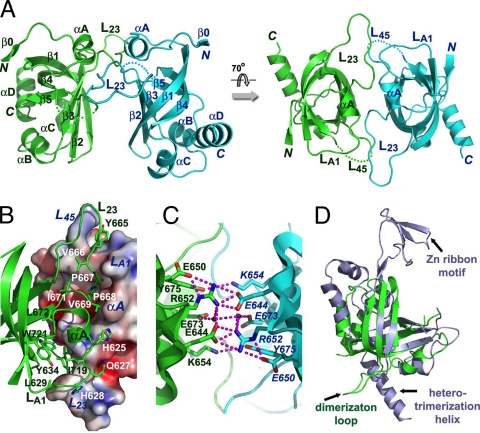
Fig 5.
Structure of the C-terminal OB fold of C. glabrata Cdc13. (A) Ribbon diagram of two views of the CgCdc13OB4 dimer. The two subunits are colored in green and cyan, respectively. The secondary structural elements are labeled. The CgCdc13OB4 dimer at right is rotated by 70° about a horizontal axis relative to the dimer at left. (B) The hydrophobic dimer interface of CgCdc13OB4. One CgCdc13OB4 molecule is in surface representation and colored according to its electrostatic potential. The other molecule is in ribbon representation and colored in green. Side chains of residues in loops LA1, L23, and L45 important for dimerization are shown as stick models. (C) An extensive electrostatic interaction network is formed by a cluster of symmetry-related charged and polar residues on the β1-β2-β3 side of the barrel. The intermolecular hydrogen bonds are shown as dashed magenta lines. (D) Superposition of CgCdc13OB4 on the crystal structure of human RPA70C reveals that CgCdc13OB4 is not structurally similar to RPA70C (3). CgCdc13OB4 and RPA70C are colored in green and light blue, respectively. The superposition is based on the OB fold β-barrels of the proteins.