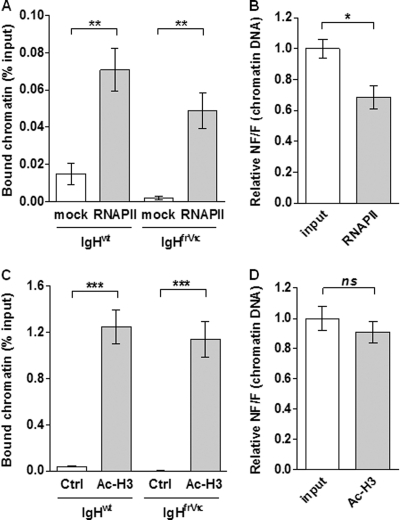
Fig 2.
Equivalent transcription and accessibility of functional and nonfunctional IgH alleles. ChIP assays were performed with spleen B cells isolated from heterozygous IgHfrVκ/wt mice and stimulated with LPS for 4 days. Immunoprecipitations were done by using antibodies to RNA polymerase II (RNAPII) (A and B) and acetylated histone H3 (Ac-H3) (C and D). (A and C) The relative enrichments (percent input) were analyzed by quantitative PCR using the allele-specific primers described in the legend of Fig. 1A (sets A and B) and were compared to those of negative controls obtained without Ab (mock) (A) or using a control IgG Ab (ctrl) (C). (B and D) Nonfunctional/functional (NF/F) ratios were obtained from these data, and the mean of ratios obtained for chromatin inputs (NF/Finput) was set to 1. Results from 5 (A and B) or 4 (C and D) independent ChIP experiments are shown. (ns [not significant], P > 0.05; ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001).