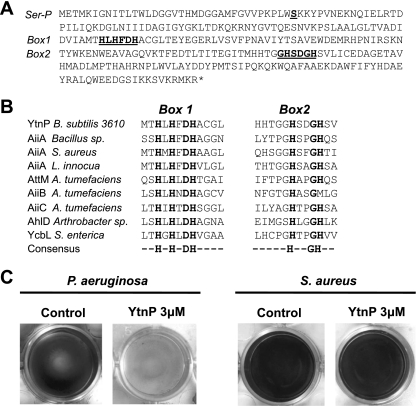
Fig 1.
YtnP is a lactonase-homologous protein. (A) Amino acid sequence of YtnP from B. subtilis strain 3610. Conserved motifs present in other metallolactamases are marked as Box 1 and Box 2 and are underlined and bold. Also an additional serine (Ser36) important for YtnP activity is marked as Ser-P. (B) Alignment of the conserved motifs of the active site of the following: YtnP and other metallolactamases; AiiA from Bacillus sp. and homologues from S. aureus and L. innocua; AttM, AiiB, and AiiC from A. tumefaciens; AhlD from Arthrobacter sp.; and glyoxalase II (YcbL) from S. enterica. The last lane of the alignment shows the consensus sequence. (C) Addition of YtnP to the biofilm formation assays inhibited biofilm formation in P. aeruginosa but not in S. aureus. Biofilm formation is observed as a pellicle attached to the submerged surfaces (at the bottom of the well plate). Crystal violet staining was used in the assay for better visualization, according to the protocol described by O'Toole and Kolter (37). For the quantification of biofilm formation, the crystal violet associated with biofilms was dissolved in acetic acid (33%) and was measured spectrophotometrically with an optical density at 595 nm (37).