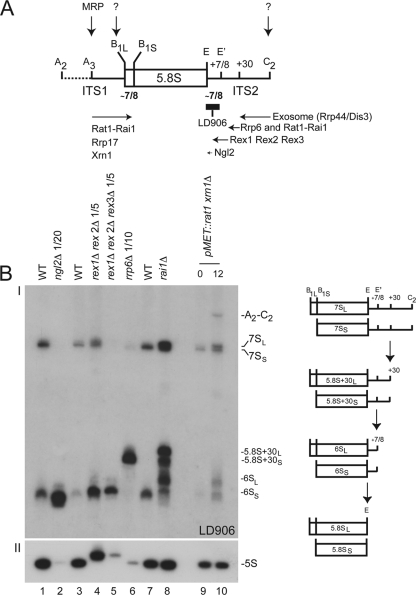
Fig 2.
How is 5.8S rRNA synthesized in budding yeast? (A) Summary cartoon. The cleavage sites (A2 to E) and trans-acting factors involved in 5.8S rRNA maturation are indicated. (B) The successive steps of 5.8S rRNA 3′-end formation are illustrated by a high-resolution Northern blot analysis of different mutants and isogenic control wild-type cells (I). Schematics depicting the structure of each physiological intermediate detected with a probe that overlaps with the 3′ end of 5.8S rRNA (LD906; see panel A) are provided to the right. Total RNA was extracted from ngl2, rai1, rex1 rex2, rex1 rex2 rex3, rrp6, and rat1 xrn1 mutants and analyzed by Northern blotting (see Materials and Methods). Ten micrograms of total RNA was loaded in each lane unless stated otherwise. As a control, panel II was hybridized with a probe specific to the 5S rRNA, as Rex1 and Rex2 have known functions in 5S rRNA 3′-end formation (46).