Fig 5.
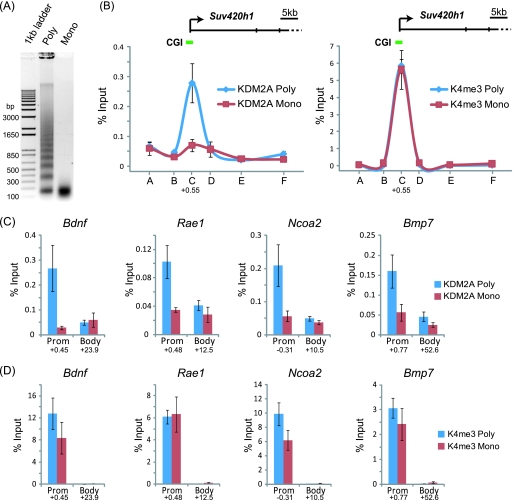
KDM2A is targeted to CpG islands through recognition of linker DNA. (A) Formaldehyde-fixed chromatin was digested with micrococcal nuclease to produce polynucleosomes that contain linker DNA and mononucleosomes lacking linker DNA. (B) KDM2A binding to the Suv420h1 gene was analyzed by ChIP at tiled positions across the gene (A to F on the x axis) by quantitative PCR on both chromatin preparations. KDM2A binding (left panel) in the polynucleosomal fraction (blue line) corresponded to the CpG island region, and this signal was lost in the mononucleosomal preparation (red line). In contrast, the signal for H3K4me3 over the same regions remained constant for both samples (right panel). (C and D) The same analysis was extended to a series of four more CpG island genes, analyzing both the CpG island region and body region. In all instances, digestion to mononucleosomes released KDM2A from CpG island chromatin (C), while the H3K4me3 signal remained relatively constant (D). Together, these observations indicate that KDM2A interacts with linker DNA at CpG islands. In all cases, error bars correspond to standard errors of the means from biological triplicates. Numbers below the data for promoter (Prom) and body primer sets indicate the position of the primer set (kb from center of amplicon) with respect to the major transcription start site of the gene in question.