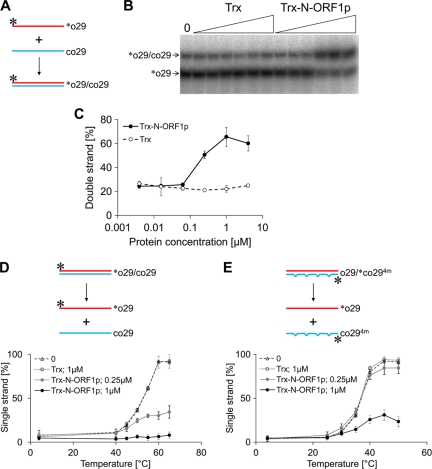
Fig 2.
The N-terminal portion of ZfL2-1 ORF1p accelerates nucleic acid annealing and keeps double-stranded DNA stable. (A) Schematic diagram of the nucleic acid annealing assay. The two complementary oligonucleotides, o29 and co29, are indicated by red and blue lines, respectively. Their sequences are shown in Table 2. o29 was labeled by 32P at its 5′ end (asterisks). (B) A representative autoradiogram of [32P]o29 separated by native PAGE. The two oligonucleotides were incubated with 4-fold serial dilutions of Trx or the Trx fusion of the N-terminal portion of ZfL2-1 ORF1p (Trx-N-ORF1p) (triangles, 0.0039 to 4 μM). The left lane indicates the result with no protein (0). (C) Acceleration of annealing. Five independent experiments were performed, and the averages and standard deviations are shown. (D and E) Schematic diagrams and the fraction of single-stranded labeled oligonucleotides in the melting assays. The 32P-labeled duplex ([32P]o29/co29 in panel D or o29/[32P]co294m in panel E) was mixed with no protein (0), 1.0 μM Trx, 0.25 μM Trx fusion of the N-terminal portion of ZfL2-1 ORF1p (Trx-N-ORF1p), or 1 μM Trx-N-ORF1p and then incubated at the indicated temperatures for 5 min. The oligonucleotide sequences are shown in Table 2. One strand of each initial duplex was labeled by 32P at its 5′ end (asterisks). Five independent experiments were performed, and the averages and standard deviations are shown.