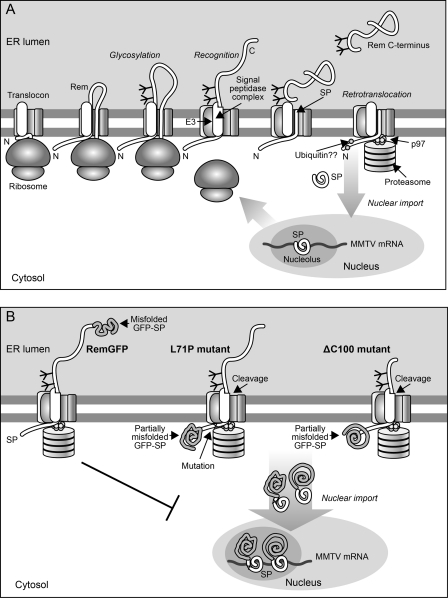
Fig 8.
Model for Rem processing. (A) Rem processing and trafficking pathway. The nascent Rem protein is recruited to the translocon by SRP to allow translation across the ER membrane. The hydrophobic domain predicted to bind SRP is adjacent to the signal peptidase cleavage site (13), suggesting that the Rem N terminus localizes to the cytoplasm. Since uncleaved Rem can be glycosylated, addition of sugars may occur prior to signal peptidase cleavage. Multiple cellular factors, including PDI proteins, likely bind to the Rem C terminus to assist glycosylation and folding. The misfolded state of Rem triggers association with the retrotranslocon, which may represent a translocon with additional components or a separate channel (25, 46). Components of the retrotranslocon include an E3 ligase, an enzyme required for the polyubiquitination of misfolded substrates (3, 34). Many substrates, including Rem, require the p97 ATPase to assist their extraction from the ER to the cytosol (7, 9, 10). Cleaved MMTV-SP then escapes proteasomal degradation to allow nuclear entry and nucleosomal targeting prior to MMTV RNA binding. The NES within MMTV-SP provides a bridge between the viral RNA and the Crm1 export protein for nuclear export of viral RNA (17, 36, 42). (B) Model for defects in Rem mutants. After Rem tagging at the C terminus, signal peptidase cannot cleave MMTV-SP and GFP does not fold properly. Failure to achieve cleavage prevents nuclear import and MMTV-SP function. In the L71 mutants with an N-terminal GFP tag, signal peptidase cleaves MMTV-SP, but a change in folding (as detected by lower fluorescence in the absence of MG132) prevents proper function after nucleolar localization. The C-terminal deletion mutants have a defect similar to the L71 mutants, except that GFP fluorescence (and folding) are further reduced, leading to defective nuclear function of MMTV-SP with a wild-type sequence.