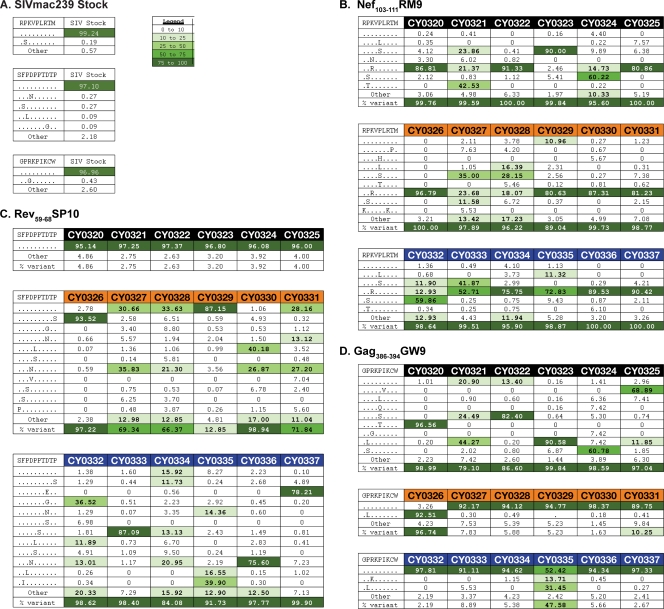
Fig 2.
Amino acid variation within three epitope sequences at 4 weeks after SIVmac239 infection. (A) The SIVmac239 inoculum was subjected to full-genome Roche/454 pyrosequencing. No significant variation was detected in the three epitope sequences Nef103-111RM9, Rev59-68SP10, and Gag386-394GW9 in the SIVmac239 inoculum. Variants were included in the table if they were also present in panel B, C, or D. (B, C, and D) Viruses replicating in the plasma samples of all 18 animals 4 weeks after SIVmac239 infection were subjected to full-genome Roche/454 pyrosequencing. Amino acid variants in the three epitopes Nef103-111RM9, Rev59-68SP10, and Gag386-394GW9 in M1/M1 animals (black), M1/M3 animals (orange), and M3/M3 animals (blue) were interrogated. Sequence variation is shown for Nef103-111RM9 (B), Rev59-68SP10 (C), and Gag386-394GW9 (D). The wild-type sequence is shown at the top of each table. Masked identical bases are shown with a dot, and amino acid replacements are shown with a capital letter. The frequency of a given epitope variant sequence is shown. A variant sequence had to be present in at least one animal at a frequency of 5% or greater to be included in the table. The legend indicates the color associated with a given variant frequency. A “0” indicates that no reads were detected with that sequence.