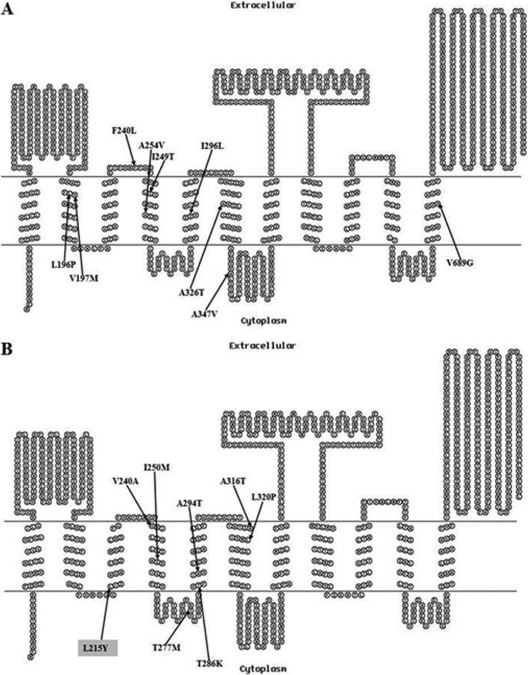
Fig 2.
Predicted topology of MmpL3 from M. smegmatis (MSMEG_0250) (A) and M. bovis BCG (BCG0243c) (B). The arrows indicate the amino acid substitutions observed for M. smegmatis (A) (see also Table 2) and M. bovis (B) (see also Table 3) mutants resistant to BM212. The amino acid substitution observed for the only resistant M. tuberculosis mutant is highlighted in gray. The MEMSAT2 program was used to predict topology (11).