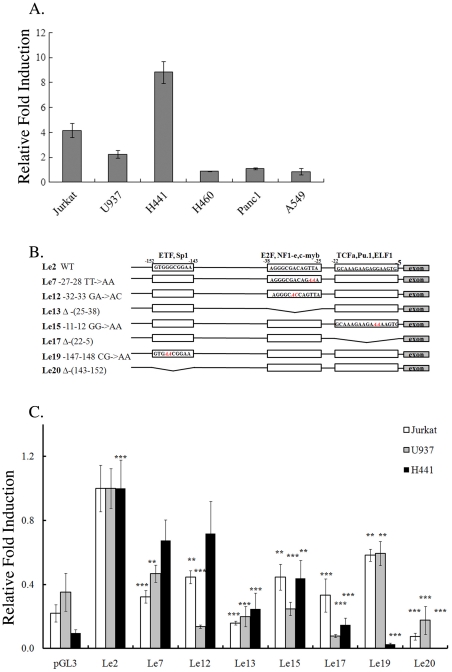
Figure 2. The vav1 5′ untranscribed sequences contain cell-type specific cis-regulatory elements.
(A) Expression of wild-type (wt) luciferase reporter gene (Le2) in cell lines from various tissue origins. Le2 was transfected into the cell lines as described in Materials and Methods and luciferase activity was measured 24 hr later. Data show luciferase activity normalized to Renilla transfection efficiency control and calculated relative to the luciferase activity of an empty vector expression, pGL3. The experiments were repeated five times. (B) Schematic map of the 5′ regulatory region of the human vav1 gene. Three putative transcription factor binding sites are highlighted by boxes. The changes introduced in these regions are as follows: nucleotide substitutions (red) and deletions (crooked lines). (C) The effect of these mutations/deletions was analyzed in Jurkat T cells, U937 myeloid cells and H441 lung cancer cells. Following transfection with plasmids containing luciferase under wt (Le2) or mutated vav1 promoter, the luciferase activity was measured and fold induction of activity was calculated relative to the activity of Le2. Experiments were repeated five times. Statistics were performed using the unpaired student T test. (**) indicates p<0.05 value and (***) indicates p<0.01.