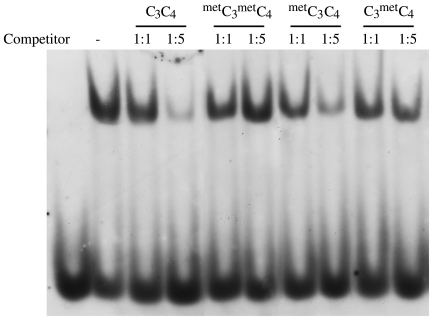
Figure 7. Methylation on CpG dinucleotides at putative transcription factor binding sites changes the affinity of protein complexes for the vav1 regulatory region.
(A) EMSA was performed with Jurkat T cell nuclear extracts and lil3-4 labeled oligonucleotide. The probe was created by annealing complementary oligonucleotides lil79 and lil80 (Table 3).-3′). The following unlabeled competitors were added: unmethylated lil79-80 oligonucleotide (C3C4); oligo methylated on both CpG methylation sites (metC3 metC4); oligo methylated only on CpG3 (metC3C4), or only on CpG4 (C3 metC4). Competitor oligonucleotide was added in an amount equal to the labeled oligo (1∶1) or in 5 molar excess (1∶5).