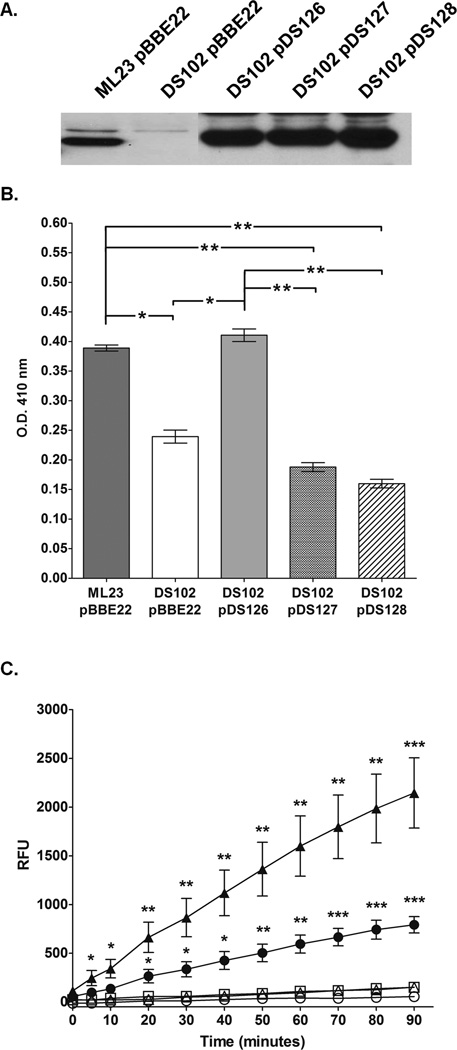
Figure 4.
Mutagenesis of the putative active site serine of BB0646 abrogates lipase activity in B. burgdorferi, but not BB0646 protein production. (A) Production of BB0646 in point mutants. The levels of BB0646 are compared between the parent (ML23/pBBE22), mutant (DS102/pBBE22), complement (DS102/pDS126), and the point mutants within the GXSXG conserved motif (DS102/pDS127; contains the bb0646-S152A allele; and DS102/pDS128; contains the bb0646-S152T allele). Whole cell equivalents were resolved by SDS-PAGE, immunoblotted and probed with antisera to BB0646. (B) Mutagenesis at serine 152 of bb0646 reduces recognition of saturated fatty acid substrate. The B. burgdorferi strains indicated above were tested for lipolytic activity against p-nitrophenyl palmitate as indicated in Fig. 3A. Note the statistically significant decrease in lipase activity for strains carrying the S152A (DS102/pDS127) and S152T (DS102/pDS128) alleles of bb0646 relative to the parent (ML23/pBBE22) and complement (DS102/pDS126). The activity observed for the bb0646 point mutants is commensurate with that observed for the bb0646 mutant (DS102/pBBE22). The single and dual asterisks denote a P value of < 0.05 and 0.01, respectively. (C) bb0646 S152 mutants do not recognize polyunsaturated fatty acid substrates. Whole cell lysates from the B. burgdorferi strains were tested for lipolytic activity against 7-HC linolenate as indicated in Fig. 3B. Samples from the parent (ML23/pBBE22; closed circles), mutant (DS102/pBBE22; open circles), the complement (DS102/pDS126; closed triangles), and the bb0646-S152 mutants (DS102/pDS127 [open triangles] and DS102/pDS128 [open squares]) were tested for their ability to cleave 7-HC linolenate over time. Cleavage of the substrate releases a fluorogenic reporter and the increased fluorescence is plotted as relative fluorescent units (RFU). Significance was measured at each time point between the bb0646 mutant and S152 mutant complements relative to the parent strain and the native complement. Bars indicate standard error. Single, dual, and tri asterisks denote P values of < 0.01, 0.001, and 0.0001 respectively.