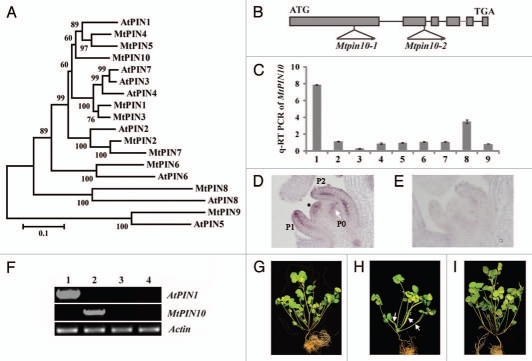
Figure 1.
Phylogenetic relationships of Medicago and Arabidopsis PINs and genetic complementation of Mtpin10. (A) Phylogenetic relationships of Medicago and Arabidopsis PINs. MtPIN4, MtPIN5 and MtPIN10 are clustered with AtPIN1. (B) MtPIN10 gene structure and Tnt1 insertion sites in Mtpin10-1 and Mtpin10-2 mutants. (C) Quantitative RT-PCR analysis of MtPIN10 expression pattern. MtPIN10 expression was normalized with an internal control, MtActin. 1, shoot bud; 2, young leaf; 3, mature leaf; 4, rachis; 5, petiole; 6, stem; 7, root; 8, flower and 9, immature pod. (D) RNA in situ hybridization. MtPIN10 transcripts were detected in the shoot apical meristem (asterisk), and P0, P1 and P2 compound leaf primordia. In P1 and P2 leaf primordia, MtPIN10 transcripts were detected in both epidermal and vascular cells. (E) No signal was detected in an adjacent tissue section hybridized with a sense probe. (F) RT-PCR analysis of AtPIN1 and MtPIN10 gene expression. Lanes 1–4, Mtpin10-1 mutant transformed with an Arabidopsis PIN1::PIN1:GFP construct, wild-type (R108), Mtpin10-1 and Mtpin10-2 mutants, respectively. MtActin was used as a loading control. (G–I) Growth defects and genetic complementation of Mtpin10 mutants. Shown were six-week-old wild type (G), Mtpin10-1 (H) and Mtpin10-1 transformed with an Arabidopsis PIN1::PIN1:GFP construct (I). In Mtpin10 mutants, leaves were often fused (H, arrows).