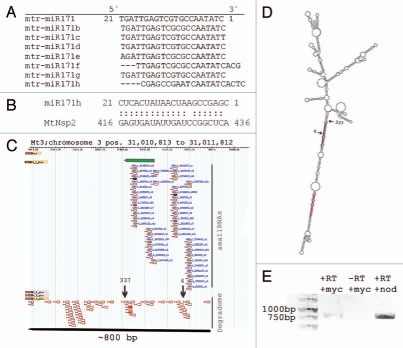
Figure 1.
Analysis of the mtr-miR171 h genomic locus. (A) Sequence alignment of all known Medicago truncatula miR171 family members. (B) Alignment of miR171 h to its cleavage target MtNsp2. (C) Distribution pattern of small RNA and degradome tags within 1 kb containing the genomic locus of the miR171 h predicted precursor. The upper short arrow indicates the predicted 109 nt long precursor. Gradient shading of the small RNA and degradome marks indicates the absolute abundance of reads at this position. (D) Secondary structure of the miR171 h primary transcript depicting the miRNA/miRNA * duplexes (bold) and DCL 1 as well as RISC cleavage sites (arrows labeled with 337 and 4, respectively). (E) Agarose gel analysis of RT PCR products validating the presence of a long primary transcript in mycorrhizal (myc) and nodulated (nod) roots. -RT terms the reverse transcription control without reverse transcriptase.