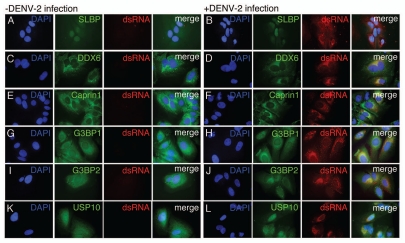
Figure 2.
DDX6, Caprin1, G3BP1, G3BP2 and USP10 colocalize with sites of DENV replication. HuH-7 cells on coverslips were infected with DENV-2 NGC at an MOI of 1.0 for 24 hours prior to fixation and probing with antibody specific for either SLBP (A and B), DDX6 (C and D), Caprin1 (E and F), G3BP1 (G and H), G3BP2 (I and J) or USP10 (K and L) and dsRNA as a marker for sites of DENV replication as previously shown by Welsch et al. The coverslips probed for SLBP and dsRNA were prepared at a different time, but under similar conditions, than the coverslips used in the rest of the localization studies described here. However, the same preparation of coverslips was also probed for DDX6 and dsRNA with the same colocalization pattern as observed with the other coverslips probed for DDX6 and dsRNA. In the final wash, DAPI was added to visualize cell nuclei and coverslips were sealed prior to visualization using an Olympus IX71 epifluorescent microscope and DP71 digital camera. Images were processed using the ImageJ software package.