Fig 2.
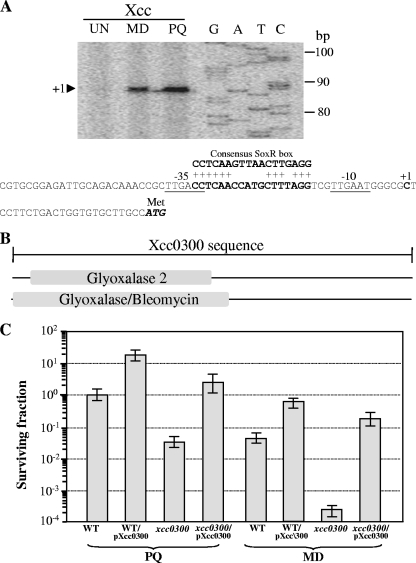
Characterization of the xcc0300 promoter and analysis of physiological roles of xcc0300. (A) Primer extension was performed to localize the 5′ ends of the xcc0300 transcripts. The primer extension products from a reaction mixture containing 32P-labeled primer BT2740 and 10 μg RNA extracted from wild-type X. campestris pv. campestris cells cultivated under uninduced (UN) or MD- or PQ-induced conditions were separated on a sequencing gel. A DNA ladder (G, A, T, and C) was prepared by using a sequencing kit with a labeled pUC/M13 forward primer and pGEM-3Zf as the template. Numbers to the left indicate DNA sizes in base pairs. An arrowhead represents the putative soxR transcription start site (position +1). The putative −10 and −35 elements of the xcc0300 promoter are underlined. The consensus E. coli SoxR-binding box is aligned above the xcc0300 promoter sequence, and the conserved residues are indicated by a plus sign. (B) Domain structure of the putative Xcc0300 protein. The 225-amino-acid sequence of Xcc0300 was analyzed by using the InterProScan algorithm (36). Glyoxalase/bleomycin represents the glyoxalase/bleomycin resistance protein/dihydroxybiphenyl dioxygenase superfamily. (C) The X. campestris pv. campestris wild-type strain harboring either the pBBR1MCS-4 vector control (WT) or pXcc0300 (WT/pXcc0300) and the xcc0300 mutant strain harboring either pBBR1MCS-4 (xcc0300) or pXcc0300 (xcc0300/pXcc0300) were grown to the exponential phase. A plate sensitivity assay was then performed by using SB agar plates containing either 1 mM PQ or 250 μM MD. The surviving fraction was calculated by dividing the number of CFU on plates containing an oxidant by the number of CFU on plates lacking an oxidant. Experiments were performed in triplicate, and the means ± standard deviations are shown.