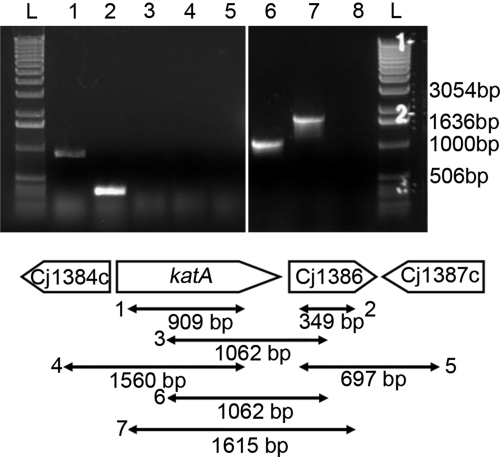
Fig 1.
The katA and Cj1386 genes are independently transcribed. RT-PCR products from RNA extracted from C. jejuni NCTC11168 were analyzed by agarose gel electrophoresis for operon identification analysis. Lanes 1 to 5, RT-PCR from RNA extracted from C. jejuni NCTC11168; lanes 6 and 7, RT-PCR from genomic DNA extracted from C. jejuni NCTC11168. Lanes: L, 1-kb ladder; 1, katA-RT-SE and katA-RT-AS (909-bp band visible, demonstrating presence of katA transcript); 2, Cj1386-RT-SE and Cj1386-RT-AS (349-bp band visible, demonstrating presence of Cj1386 transcript); 3, katA-RTint-SE and Cj1386-RTint-AS (absence of predicted 1,062-bp band, indicating that katA and Cj1386 are not cotranscribed); 4, Cj1384c-RT-SE and katA-RT-AS (absence of predicted 1,560-bp band, indicating that Cj1384c and katA are not cotranscribed); 5, Cj1386-RT-SE and Cj1387-RT-AS (absence of predicted 697-bp band, indicating that Cj1386 and Cj1387c are not cotranscribed); 6, positive-control katA-RTint-SE and Cj1386-RTint-AS (1,062-bp band present); 7, positive-control katA-RT-SE and Cj1386-RT-AS (1,615-bp band present); 8, negative control (no RT added to reaction mixture).