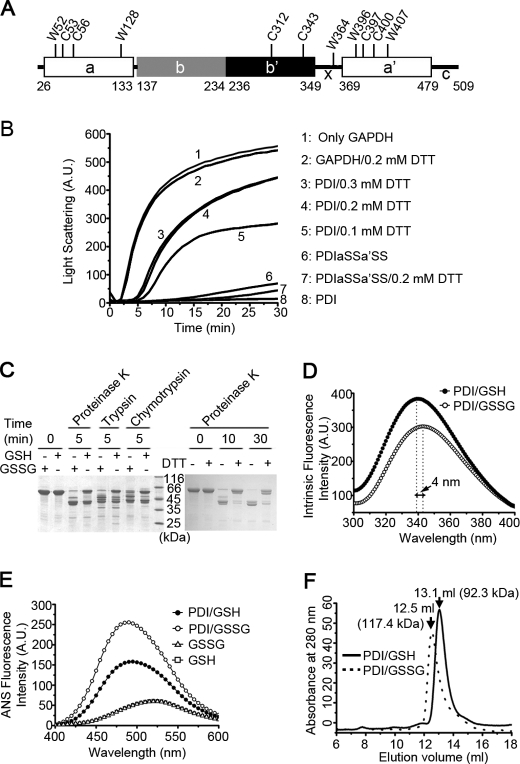
FIGURE 1.
The chaperone activity and overall conformation of hPDI are redox-regulated. A, a schematic representation of hPDI with domain organization in abb′xa′c. The positions of all of the tryptophan and cysteine residues are marked, and the residue numbering is for hPDI with signal sequence. B, hPDI is a redox-regulated chaperone. Guanidine hydrochloride-denatured GAPDH at 0.14 mm was 50-fold diluted into refolding buffer in the absence or presence of 28 μm hPDI proteins with various concentrations of DTT as indicated. Aggregation produced during the refolding was monitored by recording the light scattering at 488 nm, and the suppression of aggregation was used to measure the chaperone activity of hPDI proteins. A.U., arbitrary units. C, protease digestion profiles of hPDI at different redox states. hPDI at 1 mg/ml was incubated with 1 mm GSSG, GSH, or DTT at 25 °C for 30 min and then digested by 2 μg/ml proteinase K, trypsin, or chymotrypsin for various time as indicated. The reactions were terminated by adding PMSF to a final concentration of 0.5 mm and analyzed by reducing SDS-PAGE. The oxidized hPDI is more accessible for protease digestion than the reduced hPDI. D and E, fluorescence spectra of hPDI at different redox states. hPDI at 5 μm was incubated with 1 mm GSSG or GSH at 25 °C for 30 min, and the intrinsic (D) and ANS (E) fluorescence spectra were recorded. A.U., arbitrary units. F, size exclusion chromatography profiles of hPDI treated with 1 mm GSH (solid curve) or GSSG (dashed curve) at 25 °C for 30 min. The molecular weight of hPDI in different redox states calculated according to the elution volume is marked.