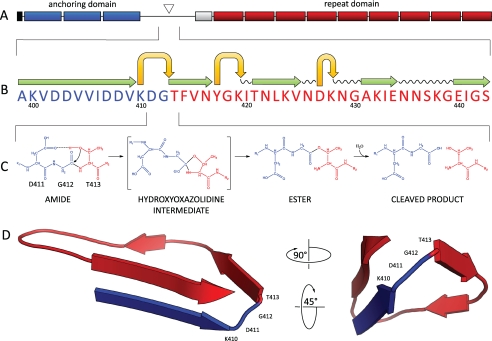
FIGURE 1.
A, domain structure of CwpV. The N terminus of CwpV contains a signal sequence (black), followed by three PF04122 cell wall anchoring motifs (blue), and a region containing the cleavage site (triangle). A serine-glycine rich region (white) precedes a number of repeats (red). B, predicted secondary structure of the region spanning the CwpV cleavage site generated using Phyre. A motif consisting of stacked β-strands (green) and tight turns (yellow) can be seen within the cleavage site, which has been previously identified and confirmed in this study by N-terminal sequencing. C, proposed mechanism of CwpV intramolecular autoprocessing. The reactive hydroxyl of Thr-413 is deprotonated by the carboxylate group of Asp-411 and launches a nucleophilic attack on the α-carbonyl carbon of Gly-412 to form a hydroxazolidine intermediate. After a proton is transferred to the leaving amino group of Thr-413, the α-carbonyl of Gly-412 is shifted to the hydroxyl of Thr-413, leading to the ester intermediate via an N-O shift. The ester is then hydrolyzed to form two peptides, the N-terminal anchoring domain and the C-terminal repeat domain. D, three-dimensional model of CwpV cleavage site (106–135 amino acids). The tertiary structure of CwpV cleavage region was predicted using Phyre with an estimated precision of 80%. A tight turn can be observed directly upstream of Thr-413 and consisting of Gly-412, Asp-411, and Lys-410.