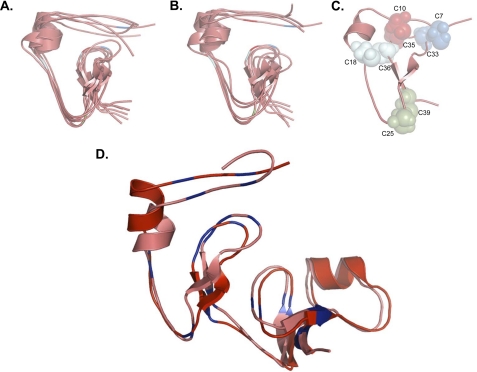
FIGURE 5.
Structural predictions and disulfide bond map of the N-terminal domain of IGFBP-5. A, de novo ab initio molecular modeling of amino acids 5 - 41 of the N-terminal domain of IGFBP-5. B, homology-based ab initio molecular modeling of amino acids 5 - 41 of the N-terminal domain of IGFBP-5. In A and B, the centers of clusters of the largest groups of highly related structures are represented by individual lines and the location of cysteine residues is indicated with the same color scheme as in C (see “Experimental Procedures” for details). C, location of disulfide bonds involving cysteines found within the conserved G32CGCCXXC39 motif based on predictions in A and B. D, de novo ab initio model of the complete IGFBP-5 N-terminal domain (amino acids 1 - 84) aligned with a model of IGFBP-5 (residues 5 - 84) based on the x-ray crystallographic structure of the N-terminal segment of IGFBP-4. Cysteines are in blue. Note that both models are nearly identical.