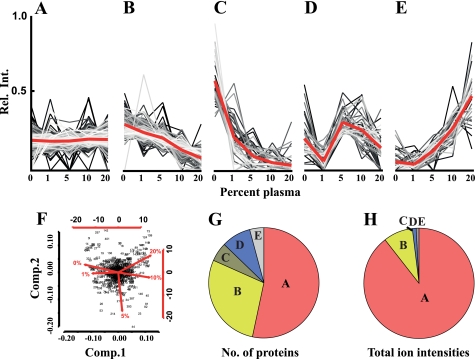
FIGURE 1.
Label-free quantitative mass spectrometry analysis of S. pyogenes adaptation to human plasma. Bacteria were grown at increasing concentrations of human plasma (0–20%), harvested, and lysed. Proteins of the lysates were identified and quantified by label-free LC-MS/MS. Using k-mean clustering the expression profiles were split into five different clusters. A, proteins that do not change in abundance. B and C, proteins with lower abundance. D and E, proteins with increased or fluctuating protein abundance levels when exposed to increasing amount of plasma. F, principal component analysis (for details, see Ref. 19) of S. pyogenes expression profiles. Each dot represents a protein and the spread of the dots and the red lines indicate the degree of proteome reorganization following exposure to increasing amounts of human plasma. G, distribution of proteins within the clusters when counting the protein from profile clusters in panel A–E. H, distribution of proteins within the clusters when summing up the ion intensities from profile clusters of panels A–E.