Background: Abnormal huntingtin (Htt)-protein interactions are implicated in Huntington disease (HD).
Results: Mutant Htt is associated with prothymosin-α (ProTα). Overexpression of ProTα protects cells against mHtt-caused toxicity, whereas knockdown of ProTα enhances the toxicity.
Conclusion: ProTα interacts with mHtt and protects against mHtt-caused toxicity.
Significance: ProTα might be a novel therapeutic target for treating HD and other polyglutamine expansion diseases.
Keywords: Aggregation, Cell Death, Huntington Disease, Polyglutamine, Protein-Protein Interactions, Prothymosin-α, huntingtin
Abstract
Huntington disease (HD), a fatal neurodegenerative disorder, is caused by a lengthening of the polyglutamine tract in the huntingtin (Htt) protein. Despite considerable effort, thus far there is no cure or treatment available for the disorder. Using the approach of tandem affinity purification we recently discovered that prothymosin-α (ProTα), a small highly acidic protein, interacts with mutant Htt (mHtt). This was confirmed by co-immunoprecipitation and a glutathione S-transferase (GST) pull-down assay. Overexpression of ProTα remarkably reduced mHtt-induced cytotoxicity in both non-neuronal and neuronal cell models expressing N-terminal mHtt fragments, whereas knockdown of ProTα expression in the cells enhanced mHtt-caused cell death. Deletion of the central acidic domain of ProTα abolished not only its interaction with mHtt but also its protective effect on mHtt-caused cytotoxicity. Additionally, overexpression of ProTα inhibited caspase-3 activation but enhanced aggregation of mHtt. Furthermore, when added to cultured cells expressing mHtt, the purified recombinant ProTα protein not only entered the cells but it also significantly suppressed the mHtt-caused cytotoxicity. Taken together, these data suggest that ProTα might be a novel therapeutic target for treating HD and other polyglutamine expansion disorders.
Introduction
Huntington disease (HD),2 a fatal neurodegenerative disorder, is caused by an expansion of polyglutamine (polyQ) in the N-terminal region of the huntingtin (Htt) protein (1). Mutation of the Htt is thought to cause gain of function, leading to progressive loss of neurons primarily in the striatum and, to a lesser extent, in the cortex (2). The age of onset of HD is usually in adulthood, with death occurring 15–20 years after disease onset (3). There is an inverse relationship between the length of the polyQ repeat and the HD onset age, such that patients with longer polyQ repeat tracts have an earlier onset age and faster progression of clinical symptoms. The clinical symptoms of HD include motor dysfuntion, psychiatric dysfunction, and cognitive decline (4). Despite considerable efforts, thus far there is no effective therapy available either to slow down or halt the progression of the disorder.
Full-length Htt is a relatively large protein (347 kDa) containing 3,144 amino acids with no sequence homology to any other known protein. After translation, full-length Htt is predominantly located in the cytoplasm, where it is cleaved by caspases and other proteases to release the small N-terminal fragments that contain the polyQ domain (5). Current evidence supports the notion that the small N-terminal fragments containing an expanded polyQ tract can not only translocate to the nucleus but also induce more potent toxicity than the full-length mutant Htt (mHtt) (6). However, how the N-terminal Htt fragments cause cytotoxicity remains poorly understood.
Protein-protein interaction plays an essential role in the regulation of almost all cell functions. When expansion of polyQ tract occurs in the Htt protein, however, the mHtt-interacting proteins are altered. Abnormal mHtt-protein interactions can, at least in part, cause HD (7). Thus, it is expected that identification and studies of mHt-interacting proteins will not only aid our understanding of the toxic mechanisms of mHtt but will also likely lead to the discovery of novel therapeutic targets for the disease. Indeed, a number of Htt-interacting proteins have been identified during the past decade (8). However, the precise roles of many of the proteins with which Htt interacts are still unclear.
ProTα is a small (12.5 kDa), highly acidic protein with a high degree of identity among mammalians that mainly resides in the nucleus, although it can also be found in the cytoplasm or even in the extracellular space (9–12). It has an unusual primary structure but lacks secondary structure (13). Although its function is still not well understood, current data suggest that ProTα facilitates cell proliferation and survival (14), remodeling of chromatin (15), and transcription (16). Previous in vitro studies have shown that ProTα is capable of preventing apoptotic cell death by inhibiting the formation of the apoptosome, a large cytosolic macromolecular complex formed in cells committed to programmed death (10). Recent in vivo studies have also revealed that ProTα not only inhibits apoptotic cell death but also suppresses necrotic cell death following ischemic stroke (17, 18). In this report, using cell culture models of HD, we demonstrate that ProTα interacts with mHtt and reduces mHtt-caused cytotoxicity.
EXPERIMENTAL PROCEDURES
Plasmid Construction
A two-step procedure was used to generate tandem affinity purification vectors expressing the N terminus of human huntingtin (Htt, 1–470 aa) containing either a normal (19Q) or expanded polyQ tract (55Q or 94Q). The N-terminal Htt sequences were first amplified from a full-length cDNA of huntingtin plasmids (kindly provided by Drs. Marian DiFiglia, Shihua Li, and Xiao-Jiang Li) by PCR using the primers forward, 5′-CACC ATG GCG ACC CTG GAA AAG CTG-3′, and reverse, 5′-GGC TGT TAA GGC AGA GCT GCT-3′. The resulting PCR products were inserted into the Gateway system entry vector, pENTR/D-TOPO (Invitrogen). To generate the destination expression vector, the Htt sequence was recombined into the pDEST/C-terminal Strep-FLAG (SF) vector via Gateway LR clonaseTM II (Invitrogen) based on the manufacturer's instructions. The generated pDEST/C-SF-Htt constructs were confirmed by DNA sequencing.
To generate the glutathione S-transferase (GST)-prothymosin-α (ProTα) recombinant protein expression construct, the full-length cDNA of ProTα was amplified from a ProTα cDNA construct (OriGene) by PCR using the primers 5′-CAG GAG CCG GAT CCA TGT CAG ACG C-3′ and 5′-TCA ATG GCG GCC GCC TAG TCA TCC TCG TCG GTC T-3′. Similarly, the inserts for the GST-tagged mutants of ProTα with deletions of different domains were generated using different primers (see supplemental data). After digestion, the resulting PCR products were inserted into the BamHI/NotI sites in the pGEX-6P-1 vector (GE Healthcare).
To generate the construct expressing GST-V5-His-ProTα recombinant protein, the following steps were used. The full-length cDNA of ProTα was first amplified from the ProTα cDNA construct by PCR using the primers 5′-CAG GAG CCG GAT CCA TGT CAG ACG C-3′ and 5′-GCT CAA TCT CGA GGT CAT CCT CGT CG-3′. After digestion with BamHI/XhoI, the resulting PCR products were inserted into pcDNA3.1/V5-His SnK/Plk2 (Addgene). V5-His-ProTα was then amplified from pcDNA3.1/V5-His-ProTα by PCR using the primers 5′-CAG GAG CCG GAT CCA TGT CAG ACG C-3′ and 5′-TCA ATG GCG GCC GCC TAC GTA GAA TC-3′. Finally, after digestion with BamHI/NotI, the resulting PCR products were inserted into the BamHI/NotI sites in the pGEX-6P-1 vector (GE Healthcare).
Generation of non-tagged ProTα and its mutants was performed by replacing the GFP inserts of a pCMV-GFP construct (purchased from Addgene) with either a full-length of ProTα or deleted ProTα cDNA after amplification by PCR using primers shown in the supplemental data and digested with EcoRI/NotI.
Generation of Stable Cell Lines Expressing C-terminal SF-tagged Htt
HEK293 cells were maintained in a complete medium containing DMEM supplemented with 10% bovine serum and 100 μg/ml of penicillin-streptomycin. When the cells were ∼70% confluent, they were transfected with pDEST/C-SF-Htt constructs using the calcium phosphate co-precipitation method (19). After 48 h, the cells were trypsinized, diluted at a 1:40 ratio, and subsequently plated onto 10-cm plates in selective medium (complete medium containing 500 μg/ml G418). The cells were grown for an additional 2–4 weeks. The medium was changed every 2 days. Individual colonies were picked with sterile 200 μl pipette tips and placed onto 12-well plates containing the selective medium. The positive clones were identified by Western blotting for expression of the bait protein. Generation of the neuronal cell lines stably expressing C-terminal SF-tagged Htt was done as described previously (20).
Tandem Affinity Purification of Interacting Protein Complex
The previously described methods were used to perform the tandem affinity purification (21). Briefly, 1 × 109 cells stably expressing the SF-tagged Htt were lysed in protein lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1mm EDTA, 1% Triton X-100 supplemented with protease inhibitor mixture (Sigma) and phosphatase inhibitor I and II (Sigma). After centrifuging for 10 min (10,000 × g) at 4 °C, the cleared supernatant was incubated for 3–4 h at 4 °C with 100 μl Strep-Tactin superflow resin (IBA). After incubation, the lysates and suspended resin were transferred into a Microspin column (GE Healthcare) and washed three times with 0.5 ml of wash buffer (100 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA). To elute the bound proteins, the resin was incubated with 0.5 ml desthiobiotin elution buffer (IBA) for 30 min. After the eluates were collected, they were transferred to another Microspin column containing 100 μl anti-FLAG M2 agarose (Sigma) and were incubated for 3–4 h at 4 °C. The beads were washed three times with 0.5 ml TBS buffer (30 mm Tris-HCl, pH 7.4, 150 mm NaCl), and then incubated for 30 min with 0.5 ml FLAG elution buffer (TBS buffer supplemented with 200 μg/ml FLAG peptide (Sigma) to elute the bound proteins).
2D-nano LC-MS/MS Analysis
The eluates obtained from the tandem affinity purification were concentrated in Microcon YM-3 protein concentration units (Millipore) and the proteins were then in-solution reduced with 50 mm DTT (Sigma) at 65 °C for 5 min and alkylated with 100 mm iodoacetamide (Sigma) before being digested with sequencing grade trypsin (Promega, Madison, WI) overnight at 37 °C. The digestion was stopped by the addition of 0.5% of acetic acid. The samples were frozen in dry ice and concentrated in a SpeedVac centrifuge (Thermo Savant). The trypsin-digested peptides were dissolved in 100 mm ammonium formate, pH 10, and separated through 2D-nanoLC with dilution using a 2D-nanoAcquity UPLC (Waters, Milford, MA). The eluted ions were analyzed by one full precursor MS scan (400–1500 m/z) followed by four MS/MS scans of the most abundant ions detected in the precursor MS scan while operating under dynamic exclusion or direct data acquisition system. Mascot server v2.3 and Mascot Daemon Toolbox v2.3 in MS/MS ion search mode (local licenses) were applied to conduct peptide matches (peptide masses and sequence tags) and protein searches against NCBInr v20100903 (11,744,690 sequences; 4,010,973,687 residues) using taxonomy filter human (for proteins identified from HEK293) or rat (for proteins identified from the rat neuronal cell lines).
Cell Culture and Transfection
HEK293 stable cell lines expressing C-terminal tagged SF-Htt containing 19Q, 55Q, or 94Q were grown in selective medium. For transfection, the different expression plasmids were introduced into HEK293 cells using a Safectine RU50 DNA transfection kit according to the manufacturer's protocol (Syd Labs).
We used previously described methods to culture and differentiate the inducible neuronal progenitor cell lines stably GFP-tagged Htt-exon1 containing 28Q or 74Q (20). For transfection, the control or the ProTα expression plasmid was introduced into the undifferentiated neurons by electroporation using a neural stem cell Nucleofector kit and device (Lonza). After transfection, the cells were cultured in differentiation medium as described previously (20) before being used for analysis.
Recombinant Protein Expression and Purification
E. coli BL21 cells which contained recombinant pGEX-6P-1::ProTα, pGEX-6P-1::V5-His-ProTα or control pGEX-6P-1 plasmids were grown with vigorous shaking in 2X YTA medium (2X Yeast Extract/Trypton medium + 100 μg/ml ampicillin) until the A600 reached 0.6 at 37 °C. 100 mm IPTG was then added to the cell cultures at a final concentration of 0.5 mm. The cells were cultured for an additional 3 h before they were transferred into centrifuge tubes and centrifuged at 7,000 × g for 10 min at 4 °C to sediment the cells. The cell pellets were stored at −80 °C.
The purification of GST, GST-ProTα, and GST-V5-His-ProTα recombinant proteins was carried out with GST fusion protein purification kit (GeneScript) according to the manufacturer's protocol. Briefly, the stored cell pellets were suspended in 20 ml ice-cold PBS buffer containing 1 mm DTT, 1 mm PMSF, and protease mixture inhibitor (1:100, Sigma), mixed gently, and sonicated. The lysates were cleared by centrifugation at 16,000 × g for 20 min at 4 °C, loaded into pre-equilibrated 1 ml GST columns, washed with 10 ml of PBS buffer, and then eluted with 4 ml of elution buffer (50 mm Tris-HCl, 10 mm reduced glutathione, pH 8.0). For further purification of V5-His-ProTα or ProTα, PreScission protease (20 units in 1 ml of PBS, GE Healthcare) was directly added to the GST column-bound GST-V5-His-ProTα or GST-ProTα and incubated at 4 °C with gentle shaking overnight. The protease digested V5-His-ProTα or ProTα was eluted with 3 ml of PBS. All protein concentrations were measured using the standard BCA assay (Thermo Scientific).
Co-immunoprecipitation Analysis and GST Pull-down Assay
Approximately, 2 × 107 cells were lysed in 1 ml of cell lysis buffer (150 mm NaCl, 1 mm EDTA, 50 mm Tris-HCl, pH 7.4, 1% Triton X-100, and protease inhibitor mixture) for 20 min at 4 °C with gentle rotation. The insoluble debris was removed by centrifugation at 10,000 × g for 15 min. For co-immunoprecipitation, cell lysates (about 1.2 mg protein) were incubated with 100 μl of Strep Tactin matrix (IBA) at 4 °C overnight with gentle rotation. The beads were washed three times with washing buffer (100 mm Tris-HCl, pH 8.0, 150 ml NaCl, 1 mm EDTA) containing 0.5% Triton X-100 and eluted by boiling at 100 °C for 5 min in 80 μl of SDS protein sample buffer. Alternatively, cell lysates were incubated with 2.4 μg of ProTα antibody (Santa Cruz Biotechnology) for 2 h at 4 °C with gentle rotation, and then the ProTα-Ab mixtures were incubated with 100 μl of pre-cleared Protein G Plus-agarose beads (Santa Cruz Biotechnology). The beads were then washed three times with PBS buffer containing 0.5% Triton X-100 and eluted with 80 μl of 2× SDS sample buffer at 100 °C for 5 min.
For the GST pull-down assay, a total of 600 μg of protein derived from the bacterial lysates expressing GST or GST-ProTα was loaded onto pre-equilibrated GST beads (GeneScript) and incubated at 4 °C for 2–3 h with gentle rotation followed by spinning at 550 × g to remove the supernatant. Next, the supernatant containing ∼650 μg of total proteins derived from the HEK293 cells expressing Htt-19Q, 55Q, or 94Q were incubated with the GST beads binding with GST or GST-ProTα at 4 °C for an additional 2–3 h with gentle rotation followed by spinning at 550 × g to remove the supernatant. The beads were washed three times with PBS buffer containing 0.5% Triton X-100 and eluted by boiling at 100 °C for 5 min in 80 μl of SDS protein loading buffer.
Immunocytochemical Staining
Immunocytochemical staining was performed according to our previously described method (20). Briefly, cells were fixed in 4% paraformaldehyde in PBS for 25 min, permeabilized with 0.15% Triton X-100 for 12 min, and incubated in blocking buffer (5% BSA in PBS) for 45 min at room temperature. The cells were then incubated in rabbit anti-ProTα (1:200, Santa Cruz Biotechnology) or goat anti-Htt (1:100, Santa Cruz Biotechnology) together with mouse anti-V5 (1:200, Invitrogen) or mouse anti-FLAG (1:250, Sigma) for 2 h, followed by incubation with Cy3-conjugated goat anti-rabbit antibody (1:200; Jackson ImmunoResearch) or with Cy3-conjugated donkey anti-goat antibody together with Cy2-conjugated goat anti-mouse antibody (1:200; Jackson ImmunoResearch) for 45 min at room temperature. The nuclei were counterstained by DAPI (Sigma). Immunostained cells were observed with a Zeiss Axiovert fluorescence microscope and images were captured with a digital camera and Axiovision software.
Western Blot Analysis
The cells prepared for Western blot analysis were washed twice with ice-cold PBS and then lysed with a cell lysis buffer (50 mm Tris-HCl, pH 6.8, 150 mm NaCl, 20 mm EDTA, 1 mm EGTA, 0.5% SDS, 0.5% Nonidet P-40, 0.5% Sarkosyl) supplemented with fresh protease inhibitor mixture (Sigma) for 20 min on ice with intermittent agitation. Protein extracts were resolved on 10–15% SDS-PAGE gels and transferred onto nitrocellulose membranes (Whatman GmbH) either under standard transfer conditions or in 20 mm sodium acetate buffer, pH 5.0, followed by fixation with 0.5% glutaraldehyde (12, 22). The membrane was blocked with 5% nonfat milk in 1× Tris buffered saline supplemented with 0.5% Tween-20 and probed with rabbit anti-ProTα (1:500, Santa Cruz Biotechnology), mouse anti-FLAG (1:1,000, Sigma), mouse anti-V5 (Invitrogen, 1:2000), mouse anti-GFP (Syd Lab), goat anti-actin (1:500, Upstate), or mouse anti-GST (1:4,000, Abcam). Detection was performed using HRP-conjugated anti-rabbit, -mouse, or -goat antibody (Santa Cruz Biotechnology) and Super-Signal West Pico substrate (Pierce). Western blot bands were scanned and quantified by measuring pixel density using a digitizing system (UN-Scan-it gel, version 6.1).
Cell Death Assay
Cell death analysis was performed with either an MTT assay using a TACSTM assay kit (Trevigen) or the Trypan Blue exclusion method using a CountessTM automated cell counter (Invitrogen) according to the manufacturers' protocols. For differentiated neurons, nuclear fragmentation was used as an indicator of cell death in some cases by following the previously described methods (20).
RNAi Knockdown Studies
The stable cell line expressing SF-Htt94Q was plated in 6-well plates. Twenty-four hours after plating, cells were transfected with either a control or a ProTα TriFECTA RNAi kit (Integrated DNA Technology) using Lipofectamine ™ RNAiMAX reagent (Invitrogen) according to the suggested protocol by the company. The cells were allowed to grow for 2 days before the cells were treated with 100 or 200 μm H2O2 and analyzed for cell death.
Fluorogenic Caspase-3 Assay
Caspase-3-like activity was measured using a previously described method (23). Briefly, SF-Htt94Q cell lines were cultured in 12-well plates in cell growth medium. Thirty-six hours after transfection with an empty vector or ProTα plasmid, the cells were cultured in serum-free medium supplemented with 200 μm H2O2 for 12 h. The cell lysates were then collected and incubated with the fluorogenic substrate DEVD-AFC (Asp-Glu-Val-Asp-amino-4-trifluoromethyl coumarin, Santa Cruz Biotechnology Inc.) for 1 h at 37 °C in a 96-well plate. The cleaved free AFC was measured using a fluorescence multi-well plate reader (Biotek) with an excitation at 400 nm and emission at 505 nm.
Statistical Analysis
One-way analysis of variance was used for statistical analysis of the experimental results and t-tests were used for comparisons between two different groups. p < 0.05 was regarded as statistically significant.
RESULTS
Identification of ProTα as a Protein Associated with mHtt
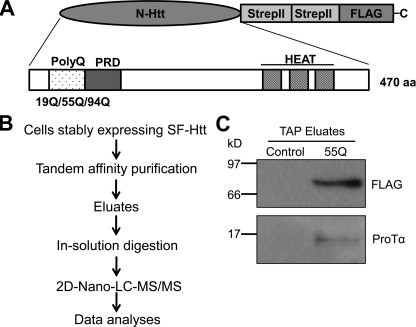
To understand the toxic mechanisms of mHtt, we recently employed the approach of tandem affinity purification (TAP) to identify novel mHtt-interacting proteins. This technique allows efficient identification of interacting proteins under near-physiological conditions (21, 24). Because the N-terminal fragment of Htt containing an expanded polyQ repeat is sufficient to cause neurotoxicity in animal and cell models (25, 26), we elected to introduce the N-terminal Htt fragment (470 amino acids) containing either 19Q (wild type), 55Q, or 94Q (mutant) into the pDEST/C-SF vector containing Strep/FLAG (SF) tandem tags at the C terminus of the insert. In addition to a polyQ region, this Htt fragment also contains a proline-rich region (PRD) and 3 HEAT repeat regions (Fig. 1A). These regions were included because mounting data suggest that most Htt interaction proteins target the N-terminal region of Htt containing not only the polyQ region but also containing the PRD and HEAT repeat regions (27, 28). To obtain sufficient expression of the SF-Htt fusion proteins for the TAP assay, we generated stable HC2S2 neuronal progenitor cells and HEK293 cell lines expressing the fusion proteins using approaches described previously (20). The neuronal cell line stably expressing SF-Htt containing 55Q was tried first for the TAP assay. After incubation of the supernatant of the neuronal cell lysates with both Strep and FLAG superflow resin columns, the final TAP eluates containing the SF-tagged Htt and associated proteins were subjected to two-dimensional nano-liquid chromatography tandem mass spectrometry (2D-nano-LC-MS/MS) (Fig. 1B). The database search of the mass spectrometry results indicated that a total of 26 proteins were identified to be associated with the SF-mHtt protein and one of them was ProTα that represented 4.35% of the bound proteins. The interaction of ProTα with mHtt was confirmed by Western blot analysis of the eluates from the TAP assay (Fig. 1C).
FIGURE 1.
Identification of ProTα as an Htt interacting protein. A, schematic illustration of the SF-Htt construct. The N-terminal fragment of Htt including the first 470 amino acids containing either 19Q (wtHtt or Htt), 55Q, or 94Q (mHtt) was introduced into a pDEST/C-SF vector containing two tandem Strep II tags and one FLAG tag at the C terminus. B, flow chart showing the experimental procedure for the identification of SF-Htt-interacting proteins. C, Western blot analysis of the proteins eluted from the TAP columns with a ProTα antibody. Control: the proteins derived from the HC2S2 cell lysate expressing an empty vector. 55Q: the proteins derived from the HC2S2 cell lysate expressing the SF-mHtt55Q fusion protein.
ProTα Interacts with mHtt
To confirm the interaction between ProTα and mHtt, we immunoprecipitated the SF-Htt fusion proteins from the cell lysates derived from the HEK293 cell lines stably expressing SF-Htt containing either 19Q, 55Q, or 94Q with Strep Tactin matrix and examined the precipitates for co-precipitation of ProTα by immunoblotting. As seen in Fig. 2A (upper panel), more ProTα was co-precipitated with SF-mHtt (55Q and 94Q) than the SF-Htt19Q protein. Similarly, when we used the approach to immunoprecipitate ProTα from the cell lysates and examined the precipitates for co-immunoprecipitation of SF-tagged Htt, we detected more SF-mHtt (55Q and 94Q) (Fig. 2B, upper panel) was co-immunoprecipitated with ProTα than the SF-Htt19Q protein.
FIGURE 2.
ProTα interacts with mHtt. A and B, ProTα and mHtt co-immunoprecipitate. The supernatants derived from cell lysates of HEK293 cells expressing the SF-tagged 19Q, 55Q, or 94Q were incubated with Strep Tactin Superflow resin (A) or protein G PLUS-agarose beads that had been coupled to a ProTα antibody (B). After centrifugation, the bound proteins were analyzed by SDS-PAGE and immunoblotted (IB) for the protein indicated on the right. C, GST pull-down assay confirming the interaction of mHtt with ProTα. Cell lysates from the HEK293 cells expressing the SF-tagged 19Q, 55Q, or 94Q constructs were incubated with glutathione resin immobilized with GST or GST-ProTα recombinant protein. The eluates from the glutathione column were analyzed by SDS-PAGE and immunoblotted for Htt (top panel, anti-FLAG) or GST (middle panel). Bottom panel: cell lysate inputs were immunoblotted with FLAG or an actin antibody.
Additionally, the interaction between the SF-mHtt and ProTα was confirmed by a GST pull-down assay. As shown in Fig. 2C, when the cell lysates derived from the HEK293 cells expressing SF-Htt19Q, 55Q, or 94Q were incubated with glutathione resin with immobilized GST or a GST-ProTα recombinant protein, the GST-ProTα fusion protein showed differential binding to the SF-Htt fusion proteins containing different polyQ repeats, with stronger binding to the mHtt (SF-Htt55Q and SF-Htt94Q) fusion protein than to the wild-type Htt (SF-Htt19Q) (Fig. 2C). These results suggest that ProTα interacts preferentially with the Htt proteins containing an expanded polyQ repeat rather than with the Htt containing the normal polyQ repeat tract.
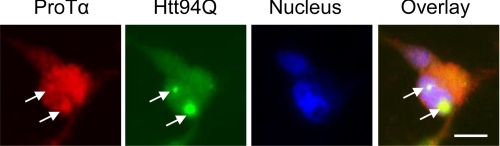
ProTα Is Colocalized with mHtt
Because ProTα is a mainly nuclear localized protein and the N-terminal proteolytic fragments of mHtt containing the polyQ repeat can also be found in the nucleus (29), we speculated that the two proteins should be colocalized in the nucleus if they interact with each other. We examined this using double immunofluorescence microscopy in the HEK293 cell lines stably expressing the SF- Htt94Q protein. As shown in Fig. 3, endogenous ProTα staining was well colocalized with both mHtt aggregates (pointed by the arrows in Fig. 3) and the soluble mHtt protein in the nucleus and cytoplasm. These data confirm that ProTα interacts with mHtt protein.
FIGURE 3.
ProTα colocalizes with mHtt. HEK293 cells stably expressing SF-Htt94Q fusion protein were fixed and stained for endogenous ProTα (red) and mHtt (green). Images were taken using a fluorescence microscope. Nuclei were stained with DAPI. Scale bar, 20 μm. Arrows point to the aggregates.
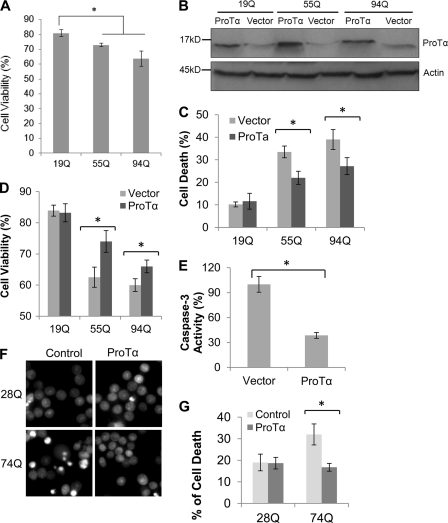
Overexpression of ProTα Inhibits mHtt-caused Cell Death and Activation of Caspase-3
In initial studies, we established that the HEK293 cells stably expressing SF-Htt55Q or SF-Htt94Q were more sensitive to exposure of 200 μm H2O2 than the cells expressing SF-Htt19Q (Fig. 4A), suggesting that expression of the SF-Htt protein containing either 55Q or 94Q sensitized cells to oxidative stress. To examine the role of ProTα in HD, we transiently transfected the HEK 293 cell lines stably expressing SF-Htt containing different polyQ repeats with ProTα expression construct (Fig. 4B). After 2 days of serum deprivation, the cells expressing the expanded polyQ proteins (55Q and 94Q) showed a significant increase in cell death compared with the cells expressing the normal polyQ repeat (19Q) Htt protein (Fig. 4, A and C). However, when the cells were transfected with ProTα significant reduction of cell death was observed in the 55Q- and 94Q-expressing cell lines (Fig. 4C). These results were confirmed by an MTT assay that assesses cell viability. As shown in Fig. 4D, overexpression of ProTα promoted cell survival in the cells expressing SF-mHtt compared with the same cell lines transfected with the empty vector.
FIGURE 4.
Overexpression of ProTα suppresses mHtt-caused cell death and caspase-3 activation. A, graph showing percentage of cell viability after the HEK293 cells stably expressing Htt19Q, 55Q, or 94Q were exposed to 200 μm H2O2 in serum-free medium for 2 days. Percentage of cell viability was calculated by determining the ratio of Trypan blue-negatively stained cells to the total cells. Data are shown as mean ± S.D.; n = 3; asterisk, p < 0.01. B, Western blot showing the expression of the three HEK293 cell lines transfected with either an expression vector expressing ProTα or with an empty vector (Vector). An equal amount of protein from each cell line was loaded on the gel. Actin is shown as a loading control. C and D, cells stably expressing SF-tagged 19Q, 55Q, or 94Q were transfected either with an empty control vector or a ProTα expression vector and then grown in serum-free medium supplemented with 200 μm H2O2 to facilitate cell death. After 48 h, cell death was analyzed by the Trypan blue exclusion method (C) and cell viability was determined by an MTT assay (D). Data are shown as mean ± S.D.; in C, a total of over 105 cells were counted; n = 4 in C; n = 3 in D; asterisk, p < 0.05. E, overexpression of ProTα inhibits caspase-3 activity. Cells expressing SF-Htt94Q were transfected with either an empty (Vector) or ProTα expression vector. Thirty-six hours after transfection, the cell growth medium was replaced with a serum-free medium supplemented with 200 μm H2O2, and cell culture was continued for 12 h before the cell lysates were collected for a caspase-3 activity assay. Data are shown as mean ± S.D.; n = 3; asterisk, p < 0.05. F, overexpression of ProTα inhibits mHtt-induced cell death in neuronal cell lines stably expressing Htt. The inducible neuronal cell lines stably expressing GFP-tagged Htt exon 1 fusion protein containing either 28Q or 74Q were then transfected with either an empty control vector (Control) or a ProTα expression vector and then grown in a neuronal differentiation medium for 3 days. The cells were then treated with 50 μm H2O2 for 20 h, and cell death was analyzed by staining the nuclei with Hoechst 33342. Cells with condensed/fragmented nuclei, indicative of dying cells, show bright fluorescence. G, graph showing quantification of fragmented/condensed nuclei in the experiments described in F. Data are shown as mean ± S.D.; n = 3; asterisk, p < 0.001.
Because previous data indicate that ProTα inhibits the formation of the apoptosome (10), we also examined whether ProTα suppresses mHtt-caused apoptotic cell death. As shown in Fig. 4E, overexpression of ProTα inhibits activation of caspase-3 in the 94Q cell lines. These data suggest that ProTα suppresses expanded polyQ protein-caused cytotoxicity at least partially via inhibiting caspase-3 activation and the apoptotic cell death pathway.
To further examine whether the modulation of polyQ toxicity by ProTα also occurred in neuronal cells, we transfected the ProTα expression plasmid into an inducible neuronal progenitor cell model of HD that we recently established (20). Similar to the results obtained with the HEK 293 cell lines, overexpression of ProTα also reduced expanded polyQ protein-induced cell death in neuronal cells (Fig. 4, F and G).
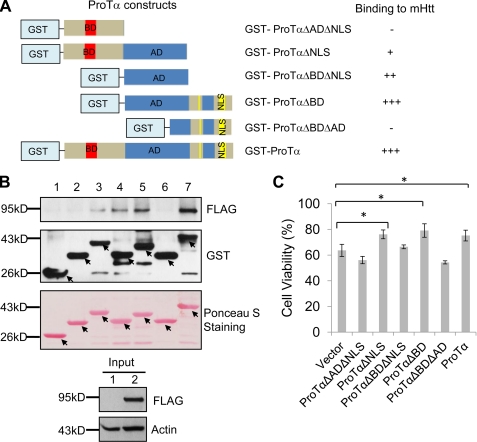
The Acidic Domain of ProTα Is Required for Its Interaction with mHtt and for Its Protective Effect against mHtt-caused Toxicity
To further examine the mHtt-binding site in ProTα, we also made GST-tagged ProTα mutants with systematic N- and C-terminal deletion of different domains of the ProTα polypeptide (Fig. 5A) and conducted additional GST pull-down experiments to examine whether GST fusions of different regions of ProTα bind SF-Htt94Q protein. The fusion proteins containing the acidic domain (between amino acids 41 and 83) bound SF-Htt94Q protein, whereas those that lacked the domain failed to bind the mHtt (Fig. 5, A and B). These results indicate that that the central acidic domain of ProTα is required for its interaction with mHtt.
FIGURE 5.
The acidic domain of ProTα is required for its interaction with mHtt and for its protective effect against mHtt-caused cytotoxicity. A, schematic drawings of the GST constructs and a summary of their binding (+, binding; −, no binding). B, GST pull-down experiment. The cell lysate expressing SF-Htt94Q was incubated with the glutathione resin immobilized with GST, GST-ProTα, or different GST-tagged mutants of ProTα as indicated. The elutes from the glutathione columns were blotted with an anti-FLAG (top panel) or anti-GST antibody (middle panel) after they attained the nitrocellulose membrane containing the GST-tagged proteins with Ponceau S (bottom panel). Arrows point to GST, GST-ProTα, or GST-tagged mutants of ProTα. 1, input from the supernatant of the HEK293 cell lysate; 2, input from the supernatant of the HEK293 cell lysate expressing SF-Htt94Q. C, acidic domain of ProTα is required for its protective effect against mHtt-caused cytotoxicity. The cells stably expressing SF-Htt94Q were transfected with the indicated constructs for 12 h and then were subjected to serum-free medium supplemented with 200 μm H2O2 for 37 h before cell viability was measured by MTT assay. Data are shown as mean ± S.D.; n = 3; asterisk, p < 0.05.
To test whether the interaction of ProTα with mHtt is required for its protective effect against mHtt-caused cytotoxicity, ProTα mutants deficient in different domains (Fig. 5, A and B) were transfected into the cell line stably expressing SF-Htt94Q and then cell viability was assessed. When the central acidic domain is absent (ProTαΔADΔNLS and ProTαΔBDΔAD), the protective effect of ProTα was abolished (Fig. 5C), whereas deletion of N-terminal basic domain (ProTαΔBD) or the C-terminal nuclear localization sequence (ProTαΔNLS) did not affect its anti-mHtt toxic effect (Fig. 5C). However, overexpression of the acidic domain polypeptide by itself did not protect SF-Htt94Q-caused toxicity (Fig. 5C), although it interacted with the mHtt (Fig. 5, A and B). These data indicate that the acidic domain is required for its interaction with mHtt and for protection against mHtt-caused toxicity. However, the domain itself may be insufficient for exerting its protective effect again mHtt-caused cytotoxicity.
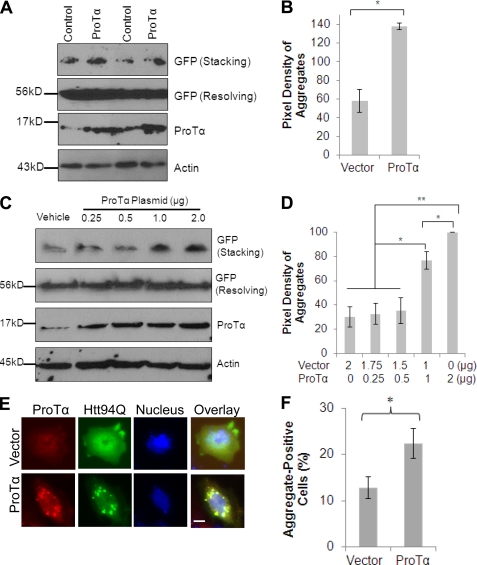
Overexpression of ProTα Enhances the Aggregation of Expanded PolyQ Proteins
As mentioned above, our previously generated HeLa cell line stably expressing GFP-74Q shows apparent protein aggregates in both the nucleus and cytoplasm (19, 30). Therefore we elected to use it to examine the effect of overexpression of ProTα on mHtt aggregation. The SDS-insoluble aggregates were seen in the stacking gel by immunoblotting (Fig. 6A, upper panel). Overexpression of ProTα in the cells increased the formation of aggregates when compared with cells transfected with an empty control vector (Fig. 6, A and B). To confirm this, the cell line expressing the GFP-Htt74Q fusion protein was transfected with an increasing amount of ProTα plasmid. As shown in Fig. 6, C and D, increased expression of ProTα was associated with enhanced aggregation of the GFP-74Q protein seen in the stacking gel. We also examined mHtt aggregates immunocytochemically in human neuron-committed NT2 cell line by co-transfection of the cell with the Htt94Q plasmid in the presence or absence of ProTα. Cell counting indicated that after exposed oxidative stress condition, more cells contained microscopically detectable Htt aggregates when transfected with ProTα rather than with the control vector (Fig. 6, E and F). These results indicate that after binding to the mHtt protein, ProTα promotes the formation of aggregates.
FIGURE 6.
Overexpression of ProTα enhances mHtt aggregation. A, HeLa cell line stably expressing GFP-tagged Htt74Q was transfected with either control or ProTα plasmid. Twenty-four hours after the transfection, the cells were subjected to SDS-PAGE and Western blot analysis for the proteins indicated on the right of each panel. B, quantitative evaluation of the Western blot Htt aggregates in Fig. 6A (mean ± S.D., n = 3, p < 0.001) showing that ProTα enhanced aggregation of mHtt. C, high levels of ProTα expression are associated with enhanced aggregation of GFP-Htt74Q fusion protein. The cells stably expressing GFP-Htt74Q were transfected with an increasing amount of ProTα plasmid as indicated above each lane. Twenty-four hours after the transfection, the cells were grown in a serum-free medium supplemented with 200 μm H2O2 for 12 h before the cells were collected for Western blot analysis. D, quantitative evaluation of aggregates in Fig. 6C (mean ± S.D., n = 3, *, p < 0.01; **, p < 0.001). E, representative images showing that ProTα enhances mHtt aggregation in neuron-committed NT2 cells. NT2 cell line was co-transfected with the SF-Htt94Q plasmid along with a ProTα or a control vector. Twenty-four hours after the transfection, the cells were grown in a serum-free medium supplemented with 200 μm H2O2 for 2 h before the cells were fixed and stained for ProTα (red) and mHtt (green). Images were taken using a fluorescence microscope. Nuclei were stained with DAPI. Scale bar, 10 μm. F, graph showing quantification of the aggregate-positive NT2 cells in the experiment described in E. Data are shown as mean ± S.D., n = 4, *, p < 0.005.
Knockdown of ProTα Levels Facilitates mHtt-caused Cytotoxicity
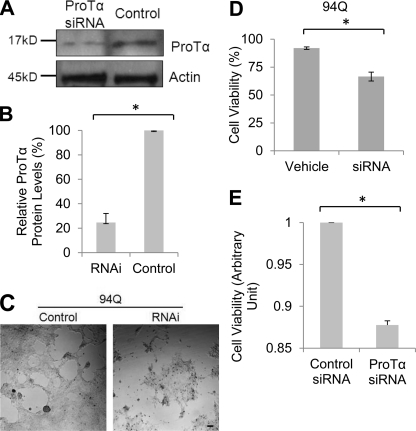
We next examined what effect reducing the levels of endogenous ProTα protein has on cell survival of the SF-Htt94Q-expressing cell line. Two days after siRNA transfection, ProTα protein levels were reduced by 70% compared with control siRNA transfected cells (Fig. 7, A and B). The cells transfected with ProTα siRNAs showed reduced proliferation, compared with the cells transfected with control siRNA (Fig. 7C). Moreover, 2 days after ProTα siRNA transfection, we observed increased cell death in the cells transfected with ProTα siRNA as compared with the cells transfected control siRNA (Fig. 7D). To examine whether the enhanced polyQ toxicity by down-regulation of ProTα also occurred in neuronal cells, we transfected a control or ProTα-specific siRNAs into an inducible neuronal progenitor cell model of HD (20). Compared with the control RNAi, knockdown of ProTα significantly enhanced GFP-Htt74Q-caused toxicity, leading to reduced viability in the cells (Fig. 7E). When aggregates were examined, however, we did not see a significant change in the number of aggregates between the cells transfected with control or ProTα-specific siRNAs (data not shown). These data suggest that down-regulation of ProTα enhances mHtt-caused cytotoxicity but may not affect the aggregation of mHtt.
FIGURE 7.
Down-regulation of ProTα enhances mHtt-caused cell death. A, Western blots showing successful knockdown of ProTα protein expression by synthetic siRNAs. Cells expressing SF-Htt94Q were transfected with control or ProTα siRNAs. Cells were harvested 2 days after the transfection and equal amounts of protein were immunoblotted for ProTα. B, graph showing quantification of band intensity shown in A. Data are shown as mean ± S.D.; n = 3; asterisk, p < 0.0001. C, representative images showing that knockdown of ProTα is associated with increased cell death in SF-Htt94Q-expressing cells. SF-Htt94Q-expressing cells that were plated in 12-well plates at an equivalent density were transfected with either control or ProTα siRNAs. The phase contrast images shown were taken 2 days after siRNA transfection. D, graph showing quantification of cell death analyzed by the Trypan blue exclusion method in the SF-Htt94Q-expressing cells with ProTα knockdown. Over 1,000 cells were counted in each experiment. Data are shown as mean ± S.D.; n = 4; asterisk, p < 0.05. E, MTT assay results showing cell viability in the neuronal cells expressing GFP-tagged Htt74Q cells after down-regulation of ProTα expression. Data are shown as mean ± S.D.; asterisk, n = 3; p < 0.0001.
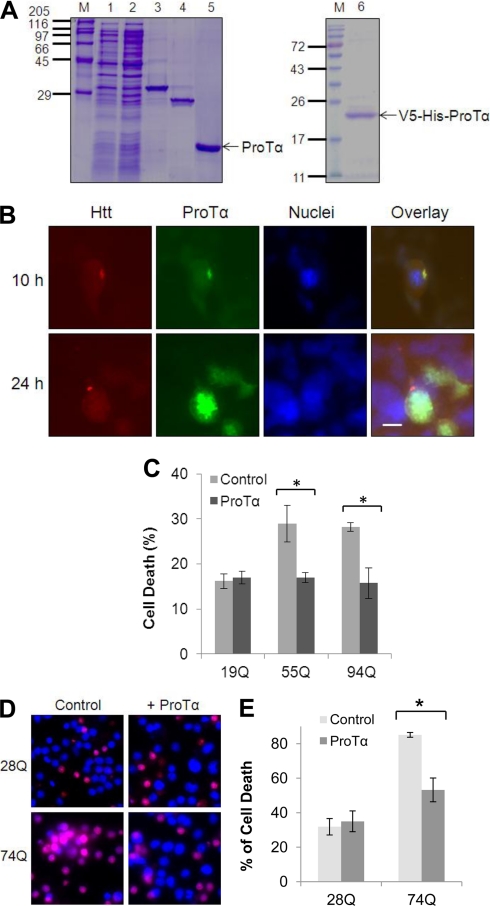
Incubation of the Cells Expressing mHtt with Purified ProTα Protein Blocks mHtt-caused Cell Death
Previous studies have indicated that addition of purified ProTα to primary neuronal cultures or treatment of ischemic animal brains with purified ProTα inhibits ischemia-caused neuronal cell death (11, 17). These observations led us to further test whether incubation of the cells expressing mHtt with purified ProTα protein can reduce the mHtt-caused cell death in cell cultures. Accordingly, we purified V5-tagged or untagged ProTα recombinant proteins using the GST affinity purification approach (Fig. 8A) and added the proteins into the cell cultures. Despite the speculation that ProTα may function as an extracellular signaling protein (11), we detected whether the uptake of the V5-tagged ProTα protein into cells occurs following its addition into the cell cultures. As shown in Fig. 8B, 10 h after the addition, V5-tagged ProTα was not only detected inside the cells but also colocalized with the mHtt. In some cells, the V5-tagged ProTα was also associated with the aggregates of mHtt (Fig. 8B, top panels). Twenty-four hours after the addition, the immunostaining of V5-tagged ProTα became relatively strong, suggesting more V5-tagged ProTα was translocated into the cells (Fig. 8B, lower panels). Moreover, addition of untagged ProTα protein into HEK 293 cells almost completely blocked cell death induced by mHtt proteins (55Q, 94Q) (Fig. 8C). Similarly, the addition of purified ProTα protein into neuronal cell cultures also remarkably inhibited GFP-tagged exon-1 Htt protein-caused toxicity (Fig. 8, D and E). These observations suggest that the ProTα protein, when applied extracellularly, can get into the cells and efficiently inhibit expanded polyQ-caused cell death.
FIGURE 8.
Addition of the purified ProTα protein to cell cultures suppresses mHtt-caused toxicity. A, purification of ProTα. GST-ProTα fusion protein was expressed in E. coli cells and purified by GST affinity method. M, protein marker; lane 1, supernatant of E. coli cell lysates containing recombinant GST-ProTα; lane 2, flow throughout after GST column binding; lane 3, GST-ProTa fusion protein; lane 4, GST protein; lane 5, Purified ProTa after GST-ProTα was digested by PreScission protease to remove GST; lane 6, Purified V5-His-ProTa after GST-V5-His-ProTα was digested by PreScission protease. B, recombinant V5-His-ProTα is co-localized with mHtt. HEK293 cells stably expressing SF-Htt94Q fusion protein were grown in normal medium supplemented with purified V5-ProTα protein (1.5 μg/ml) for 10 or 24 h. The cells were then fixed and stained for Htt (red) and V5 (green). Nuclei were stained with DAPI. Scale bar, 20 μm. C, incubation of Htt-expressing HEK293 cell lines with purified ProTα reduces cell death caused by mHtt. HEK293 cell lines stably expressing SF-tagged 19Q, 55Q, or 94Q were incubated with purified ProTα (2 μg/ml) and treated with 200 μm H2O2 for 2 days before being subjected to cell viability analysis. Data are shown as mean ± S.D.; n = 3; asterisk, p < 0.05. D, representative images showing cell death in neuronal cells. The neuronal cells stably expressing GFP-Htt28Q or GFP-Htt74Q fusion protein were differentiated in differentiation medium. Three days following neuronal differentiation, the purified ProTα was added to the cell cultures (at a final concentration of 2 μg/ml). After 24 h, the cells were treated with 100 μm H2O2 for 7 h before the cells were stained with propidium iodide (PI) or trypan blue. Purple color indicates dead or dying cells. E, graph showing quantification of cell death in the neuronal cells expressing GFP-Htt28Q or GFP-Htt74Q after addition of ProTα. Data are shown as mean ± S.D.; n = 3; asterisk, p < 0.05.
DISCUSSION
In this report, we demonstrate that ProTα protein preferentially interacts with the N-terminal mHtt proteins containing an expanded polyQ repeat. When overexpressed, ProTα reduces the expanded polyQ protein-caused cytotoxicity in both non-neuronal and neuronal cells and inhibits activation of caspase-3. In absence of the acidic domain, ProTα not only fails to interact with mHtt but loses its protective effect against mHtt-caused cytotoxicity. Additionally, overexpression of ProTα is also associated with enhanced aggregation of mHtt. In contrast, knockdown of ProTα expression by RNAi increases the expanded polyQ protein-caused cytotoxicity. Furthermore, when added to the cultured cells expressing mHtt, the purified recombinant ProTα protein not only entered the cells but also blocked the mHtt-caused cytotoxicity. These results suggest that ProTα might be novel therapeutic target for treating expanded polyQ protein-caused neurodegeneration.
Using several different approaches, we demonstrated that ProTα interacts preferentially with mHtt containing an expanded polyQ repeat but not with Htt containing normal polyQ repeats. The interaction appears to be dependent on the central acidic domain of ProTα as deletion of the domain abolishes its interaction with mHtt. Our data also suggest that the interaction of ProTα with mHtt is required but is insufficient for regulating its protective effect against mHtt-caused cytotoxicity since the ProTα polypeptide only containing the central acidic domain loses its protective effect even though it still interacts with mHtt.
Despite our observations of the acidic domain in mediating the interaction of ProTα with mHtt, it remains unclear how they interact with each other. ProTα is a natively unfolded protein that is mainly located in the nucleus (13) but can also be found in the cytosol (10) or even in the extracellular space under specific physiological or pathological conditions (11). The full-length mHtt protein containing an expanded polyQ repeat is primarily a cytoplasmic protein. However, upon cleavage, the N-terminal fragments of mHtt translocate to the nucleus and mediate their toxicity by interacting with a number of nuclear proteins. When the proteins with which the two proteins interact are compared, it is seen that both ProTα and the expanded polyQ proteins bind to CREB-binding protein (CBP) (31) and other transcription factors. However, the effects following the binding are opposite. The binding of CBP to ProTα facilitates transcription activities (31), while the binding of CBP to mHtt suppresses transcription (32). Since ProTα lacks secondary structure, it is conceivable that when the unfolded ProTα binds to the mHtt it may adopt a secondary or even a tertiary structure, which then prevents its interaction with other proteins. If the intracellular ProTα concentration is low, the free ProTα that can bind to other proteins is reduced, leading to the disruption of its anti-apoptotic (10) and anti-necrotic (17) functions and subsequently triggering cell death.
Because our data also show that overexpression of ProTα enhances aggregation of mHtt, it is possible that, upon binding to the soluble mHtt proteins, it may trigger a conformational change in the mHtt proteins, leading to enhanced aggregation. Because aggregation of the toxic mHtt is likely beneficial to cell survival (33), this may at least partially explain why the protective effect of ProTα is associated with enhanced aggregation of mHtt proteins.
Interestingly, using a primary neuronal culture model, Ueda et al. recently reported that ProTα inhibited necrotic cell death but enhanced apoptotic cell death by up-regulating proapoptotic proteins Bax and Bim but down-regulating anti-apoptotic proteins Bcl-2 and Bcl-XL when the cells were cultured at a specific density and in a serum-free medium (11). Because necrotic cell death is an overwhelming pathway by which cells die under specific cell culture condition, the overall effect of ProTα should still be protective (18). In ischemic stroke mouse studies, administration of purified ProTα remarkably inhibited both necrotic and apoptotic cell death (18). In accord with these observations, we observed that when added into the cell culture, exogenous ProTα was able to enter into the cells and colocalize with mHtt. The uptake of ProTα was associated with a remarkable protection against expanded polyQ protein-induced cytotoxicity, although we cannot exclude the possibility that ProTα may also function as an extracellular signaling protein (11). In addition, our results support the protective role of ProTα against mHtt-induced cell death by inhibiting the apoptotic cell death pathway since we observed a significant reduction of caspase-3 activation upon its overexpression of ProTα.
In summary, we show here that ProTα interacts preferentially with the mHtt protein containing an expanded polyQ repeat and inhibits its cytotoxicity by inhibiting caspase-3 activation and facilitating aggregate formation. This suggests that ProTα may be a novel therapeutic target for treating HD and other PolyQ diseases. Since ProTα is a small protein that lacks a secondary structure and can function when administrated extracellularly, it is possible that ProTα itself may be used as a therapeutic compound in treating HD and other neurodegenerative disorders.
Supplementary Material
Acknowledgments
We thank Dr. Robin Miskimins for critical reading of the manuscript; Drs. Marian DiFiglia, Shihua Li, and Xiao-Jiang Li for providing huntingtin constructs; Dr. Mervyn J. Monteiro for providing the stable cell lines expressing GFP-tagged Htt-exon-1; Justine Ferguson and Dr. Dong Zhang for assistance in cell death analysis; Dr. Khosrow Rezvani for assistance in measuring pixel density of Western blot bands; and Drs. Joyce Keifer and Fran Day at the South Dakota Imaging Core Facility (supported by NIH P20 RR 015567, which is designated as a Center of Biomedical Research Excellence [COBRE] to Dr. Joyce Keifer) for help in fluorescence microscopy.
This work was supported by Start-up Funds from the Sanford School of Medicine, University of South Dakota (to H. W.), a Competitive Research Grant Award of the South Dakota Board of Regents (to H. W.), and a New Faculty Development Award of the University of South Dakota (to H. W.).

This article contains supplemental data.
- HD
- Huntington disease
- polyQ
- polyglutamine
- Htt
- huntingtin
- mHtt
- mutant huntingtin
- ProTα
- prothymosin-α
- SF
- Strep/FLAG
- TAP
- tandem affinity purification
- AD
- acidic domain
- BD
- basic domain
- NLS
- nuclear localization sequence
- CBP
- CREB-binding protein.
REFERENCES
- 1. Bates G., Harper P., Jones L. (2002) Huntington's Disease, 3rd Ed. Oxford University Press, Oxford [Google Scholar]
- 2. Lombardi M. S., Jaspers L., Spronkmans C., Gellera C., Taroni F., Di Maria E., Donato S. D., Kaemmerer W. F. (2009) A majority of Huntington's disease patients may be treatable by individualized allele-specific RNA interference. Exp. Neurol. 217, 312–319 [DOI] [PubMed] [Google Scholar]
- 3. Vonsattel J. P., DiFiglia M. (1998) Huntington disease. J. Neuropathol. Exp. Neurol. 57, 369–384 [DOI] [PubMed] [Google Scholar]
- 4. Ross C. A., Tabrizi S. J. (2011) Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 10, 83–98 [DOI] [PubMed] [Google Scholar]
- 5. Sawa A., Nagata E., Sutcliffe S., Dulloor P., Cascio M. B., Ozeki Y., Roy S., Ross C. A., Snyder S. H. (2005) Huntingtin is cleaved by caspases in the cytoplasm and translocated to the nucleus via perinuclear sites in Huntington's disease patient lymphoblasts. Neurobiol. Dis. 20, 267–274 [DOI] [PubMed] [Google Scholar]
- 6. Orr A. L., Li S., Wang C. E., Li H., Wang J., Rong J., Xu X., Mastroberardino P. G., Greenamyre J. T., Li X. J. (2008) N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J. Neurosci. 28, 2783–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Y., Meriin A. B., Costello C. E., Sherman M. Y. (2007) Characterization of proteins associated with polyglutamine aggregates: a novel approach towards isolation of aggregates from protein conformation disorders. Prion 1, 128–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li S. H., Li X. J. (2004) Huntingtin-protein interactions and the pathogenesis of Huntington's disease. Trends Genet. 20, 146–154 [DOI] [PubMed] [Google Scholar]
- 9. Hannappel E., Huff T. (2003) The thymosins. Prothymosin alpha, parathymosin, and beta-thymosins: structure and function. Vitam. Horm. 66, 257–296 [DOI] [PubMed] [Google Scholar]
- 10. Jiang X., Kim H. E., Shu H., Zhao Y., Zhang H., Kofron J., Donnelly J., Burns D., Ng S. C., Rosenberg S., Wang X. (2003) Distinctive roles of PHAP proteins and prothymosin-alpha in a death regulatory pathway. Science 299, 223–226 [DOI] [PubMed] [Google Scholar]
- 11. Ueda H., Fujita R., Yoshida A., Matsunaga H., Ueda M. (2007) Identification of prothymosin-alpha1, the necrosis-apoptosis switch molecule in cortical neuronal cultures. J. Cell Biol. 176, 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsunaga H., Ueda H. (2010) Stress-induced non-vesicular release of prothymosin-α initiated by an interaction with S100A13, and its blockade by caspase-3 cleavage. Cell Death Differ. 17, 1760–1772 [DOI] [PubMed] [Google Scholar]
- 13. Piñeiro A., Cordero O. J., Nogueira M. (2000) Fifteen years of prothymosin alpha: contradictory past and new horizons. Peptides 21, 1433–1446 [DOI] [PubMed] [Google Scholar]
- 14. Letsas K. P., Frangou-Lazaridis M. (2006) Surfing on prothymosin alpha proliferation and anti-apoptotic properties. Neoplasma 53, 92–96 [PubMed] [Google Scholar]
- 15. Covelo G., Sarandeses C. S., Díaz-Jullien C., Freire M. (2006) Prothymosin alpha interacts with free core histones in the nucleus of dividing cells. J. Biochem. 140, 627–637 [DOI] [PubMed] [Google Scholar]
- 16. Mosoian A., Teixeira A., High A. A., Christian R. E., Hunt D. F., Shabanowitz J., Liu X., Klotman M. (2006) Novel function of prothymosin alpha as a potent inhibitor of human immunodeficiency virus type 1 gene expression in primary macrophages. J. Virol. 80, 9200–9206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fujita R., Ueda H. (2007) Prothymosin-alpha1 prevents necrosis and apoptosis following stroke. Cell Death Differ. 14, 1839–1842 [DOI] [PubMed] [Google Scholar]
- 18. Fujita R., Ueda M., Fujiwara K., Ueda H. (2009) Prothymosin-alpha plays a defensive role in retinal ischemia through necrosis and apoptosis inhibition. Cell Death Differ. 16, 349–358 [DOI] [PubMed] [Google Scholar]
- 19. Wang H., Lim P. J., Yin C., Rieckher M., Vogel B. E., Monteiro M. J. (2006) Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington's disease by ubiquilin. Hum. Mol. Genet. 15, 1025–1041 [DOI] [PubMed] [Google Scholar]
- 20. Dong G., Ferguson J. M., Duling A. J., Nicholas R. G., Zhang D., Rezvani K., Fang S., Monteiro M. J., Li S., Li X. J., Wang H. (2011) Modeling pathogenesis of Huntington's disease with inducible neuroprogenitor cells. Cell Mol. Neurobiol. 31, 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gloeckner C. J., Boldt K., Schumacher A., Ueffing M. (2009) Tandem affinity purification of protein complexes from mammalian cells by the Strep/FLAG (SF)-TAP tag. Methods Mol. Biol. 564, 359–372 [DOI] [PubMed] [Google Scholar]
- 22. Sukhacheva E. A., Evstafieva A. G., Fateeva T. V., Shakulov V. R., Efimova N. A., Karapetian R. N., Rubtsov Y. P., Vartapetian A. B. (2002) Sensing prothymosin alpha origin, mutations and conformation with monoclonal antibodies. J. Immunol. Methods 266, 185–196 [DOI] [PubMed] [Google Scholar]
- 23. Wang H., Yu S. W., Koh D. W., Lew J., Coombs C., Bowers W., Federoff H. J., Poirier G. G., Dawson T. M., Dawson V. L. (2004) Apoptosis-inducing factor substitutes for caspase executioners in NMDA-triggered excitotoxic neuronal death. J. Neurosci. 24, 10963–10973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 25. Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S. W., Bates G. P. (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87, 493–506 [DOI] [PubMed] [Google Scholar]
- 26. Ratovitski T., Gucek M., Jiang H., Chighladze E., Waldron E., D'Ambola J., Hou Z., Liang Y., Poirier M. A., Hirschhorn R. R., Graham R., Hayden M. R., Cole R. N., Ross C. A. (2009) Mutant huntingtin N-terminal fragments of specific size mediate aggregation and toxicity in neuronal cells. J. Biol. Chem. 284, 10855–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harjes P., Wanker E. E. (2003) The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem. Sci. 28, 425–433 [DOI] [PubMed] [Google Scholar]
- 28. Kaltenbach L. S., Romero E., Becklin R. R., Chettier R., Bell R., Phansalkar A., Strand A., Torcassi C., Savage J., Hurlburt A., Cha G. H., Ukani L., Chepanoske C. L., Zhen Y., Sahasrabudhe S., Olson J., Kurschner C., Ellerby L. M., Peltier J. M., Botas J., Hughes R. E. (2007) Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 3, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Atwal R. S., Xia J., Pinchev D., Taylor J., Epand R. M., Truant R. (2007) Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum. Mol. Genet. 16, 2600–2615 [DOI] [PubMed] [Google Scholar]
- 30. Wang H., Monteiro M. J. (2007) Ubiquilin interacts and enhances the degradation of expanded-polyglutamine proteins. Biochem. Biophys. Res. Commun. 360, 423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karetsou Z., Kretsovali A., Murphy C., Tsolas O., Papamarcaki T. (2002) Prothymosin alpha interacts with the CREB-binding protein and potentiates transcription. EMBO Rep. 3, 361–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Steffan J. S., Kazantsev A., Spasic-Boskovic O., Greenwald M., Zhu Y. Z., Gohler H., Wanker E. E., Bates G. P., Housman D. E., Thompson L. M. (2000) The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. U.S.A. 97, 6763–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.