summary
Objective
To ascertain a viral vector-based short hairpin RNA (shRNA) capable of reducing the interleukin-1β (IL-1β) transcript in osteoarthritis (OA)-prone chondrocytes and detect corresponding changes in the expression patterns of several critical disease mediators.
Methods
Cultured chondrocytes from 2-month-old Hartley guinea pigs were screened for reduction of the IL-1β transcript following plasmid-based delivery of U6-driven shRNA sequences. A successful plasmid/shRNA knockdown combination was identified and used to construct an adeno-associated virus serotype 5 (AAV5) vector for further evaluation. Relative real-time reverse transcription polymerase chain reaction (RTPCR) was used to quantify in vitro transcript changes of IL-1β and an additional nine genes following transduction with this targeting knockdown vector. To validate in vitro findings, this AAV5 vector was injected into one knee, while either an equivalent volume of saline vehicle (three animals) or non-targeting control vector (three animals) were injected into opposite knees. Fold differences and subsequent percent gene expression levels relative to control groups were calculated using the comparative CT (2−ΔΔCT) method.
Results
Statistically significant decreases in IL-1β expression were achieved by the targeting knockdown vector relative to both the mock-transduced control and non-targeting vector control groups in vitro. Transcript levels of anabolic transforming growth factor-β (TGF-β) were significantly increased by use of this targeting knockdown vector. Transduction with this targeting AAV5 vector also significantly decreased the transcript levels of key inflammatory cytokines [tumor necrosis factor-α (TNF-α), IL-2, IL-8, and IL-12] and catabolic agents [matrix metalloproteinase (MMP)13, MMP2, interferon-γ (IFN-γ), and inducible nitrous oxide synthase (iNOS)] relative to both mock-transduced and non-targeting vector control groups. In vivo application of this targeting knockdown vector resulted in a >50% reduction (P= 0.0045) or >90% (P= 0.0001) of the IL-1β transcript relative to vehicle-only or non-targeting vector control exposed cartilage, respectively.
Conclusions
Successful reduction of the IL-1β transcript was achieved via RNA interference (RNAi) techniques. Importantly, this alteration significantly influenced the transcript levels of several major players involved in OA pathogenesis in the direction of disease modification. Investigations to characterize additional gene expression changes influenced by targeting knockdown AAV5 vector-based diminution of the IL-1β transcript in vivo are warranted.
Keywords: Osteoarthritis, Interleukin-1β, Chondrocytes, Guinea pig, Adeno-associated virus vector, RNA interference
Introduction
Multiple molecular and biomechanical factors induce signaling cascades that lead to extracellular matrix (ECM) destruction and osteoarthritis (OA). Specifically, interleukin-1β (IL-1β) is considered a primary instigator of OA1. In cell culture and explants, IL-1β inhibits maintenance of the ECM network2 and directly provokes protease-induced cartilage destruction3. IL-1β is also a mediator of inflammation and may be involved with pain mechanisms in joints affected by OA4. Thus, it is recognized that inhibiting the biological activities of IL-1β could modify or alleviate the disease at several levels5.
In vitro and in vivo efforts to reduce or block IL-1β have been demonstrated using diacerein/rhein or IL-1 receptor antagonist protein (IRAP) in experimental OA4,6–18. Although these studies suggest that IL-1β is important for the development of secondary OA, IL-1β knockout mice show accelerated progression of OA lesions compared to wild-type litter mates1. Use of knockout mice to study the pathogenesis of OA is valuable but suffers from drawbacks, including potential for compensatory gene expression1. Alternatively, RNA interference (RNAi) technology can be used to generate loss-of-function phenotypes without complications associated with traditional knockout animals19. Targeted, localized reduction of gene products implicated in naturally-occurring OA provides the opportunity to define contributions of specific molecular pathways to the disease and offers potential as a therapeutic strategy20. Further, delivery of small RNAs capable of inciting cellular knockdown machinery via lowly immunogenic and persistent viral vectors, such as recombinant adeno-associated virus (AAV), have emerged as capable systems for application in musculoskeletal disorders21. AAV-based delivery of RNAi constructs has been successful in cell culture and animal model systems20 and holds promise in the context of OA.
Currently, there are minimal reports using RNAi to study the pathogenesis of OA. It was our hypothesis that reducing the IL-1β transcript via delivery of a plasmid-based and, subsequently, an AAV vector-based short hairpin RNA (shRNA) sequence would alter expression patterns of additional mediators implicated in OA. Importantly, this knockdown was investigated, and sustained, with and without inflammatory insults instigated by either lipopoly-saccharide (LPS) or exogenous recombinant human (rh)IL-1β. Moreover, evidence is provided that reduction of IL-1β may be plausible in vivo using this validated tool.
Materials & methods
All procedures were approved by the university's Institutional Laboratory Animal Care and Use Committee and performed in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals. Cartilage was aseptically harvested from knees of five 2-month-old male Hartley guinea pigs (Charles River Laboratories, Wilmington, MA), pooled, and minced. Cells were isolated following a 6-hour digestion in 0.2% collagenase I (Gibco, Grand Island, NY) and expanded in standard culture conditions in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 29.2 mg/ml l-glutamine, 50 U penicillin/ml, and 50 U streptomycin/ml until >95% confluent. Chondrocytes were implemented prior to their fourth passage. Six 2-month-old male animals were used for the in vivo study.
Plasmid screening
Experiments to screen plasmids containing potential knockdown shRNA sequences were performed in triplicate. The selected plasmid allowed simultaneous expression of shRNA via the U6 promoter and CMV-driven enhanced green fluorescent protein (GFP) expression [Fig. 1(A)] and is intended for making single-stranded AAV vectors. Ten shRNA knockdown sequences and one non-targeting sequence [Fig. 1(A)] were conceived using RNAi Block-IT Designer (Invitrogen, Carlsbad, CA) and cloned into plasmids using standard techniques. Chondrocytes (2.5×105) were seeded into T25 flasks (Greiner Bio-One, Monroe, NC) and allowed to reach ~50% confluency, at which time cultured cells were transfected using FuGENE®6 Transfection reagent (Roche Applied Science, Indianapolis, IN) in serum- and antibiotic-free DMEM according to recommendations at a reagent:DNA ratio of 3:1. Media containing transfection complexes were removed 24-h post-exposure; new non-supplemented Minimum Essential Media, Eagle (MEM-E) (Mediatech, Herndon, VA) was added, either with or without 1 μg/ml LPS (Escherichia coli O111:B4; Sigma–Aldrich, St. Louis, MO) for an additional 24-h. Cells were monitored for onset of GFP using fluorescent microscopy. All treatment groups were removed from culture flasks 48-h post-transfection for flow cytometry. Once a successful plasmid/shRNA knockdown sequence combination was identified, culture experiments to detect additional transcript changes were performed in triplicate.
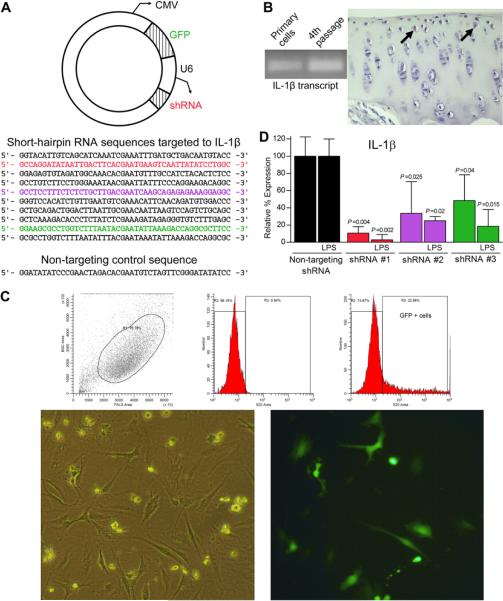
Fig. 1.
(A) Schematic representation of the AAV plasmid (top) used to deliver selected shRNA sequences (bottom) to cultured guinea pig chondrocytes. Sequences that significantly reduced IL-1β transcript expression in chondrocytes as detected by real-time RT-PCR are shown in corresponding colors in (D). (B) RT-PCR products in agarose gel (left) and positive immunohistochemistry staining (right, arrows) confirm expression of IL-1β at the transcript and protein levels, respectively, in 2-month-old primary cultured chondrocytes up to the fourth passage and in situ chondrocytes. (C) Representative example of flow cytometry data (top graphs) to confirm and sort GFP + chondrocytes (bottom photomicrographs,200× final magnification) in plasmid-transfected samples. (D) Significant reduction of the IL-1β transcript (mean percent gene expression levels ±95% confidence intervals) relative to a non-targeting control shRNA was detected in guinea pig chondrocytes receiving the color matched shRNA sequences in (A) delivered via plasmid. Within each media condition (control or LPS), fold differences and subsequent percent gene expression levels relative to C were calculated using the comparative CT (2−ΔΔCT) method. Data (triplicate assay values for triplicate culture samples) were analyzed using one-way ANOVA for comparison among groups followed by pairwise comparisons using Tukey 95% confidence intervals; significant differences are indicated.
In vitro knockdown
The selected plasmid/shRNA knockdown sequence combination (shRNA#1) and non-targeting control plasmid were used to produce IL-1β knockdown and non-targeting control AAV serotype 5 (AAV5) vectors, respectively, by the Viral Vector Core at Nationwide Children's Hospital (Columbus, OH) using the standard triple-transfection method22. Vector titration to determine DNase-resistant particles (DRP) of each vector was performed as described using quantitative real-time PCR23. Chondrocytes were seeded and cultured, as above, until ~75% confluent, at which time triplicate culture replicates were exposed to one of three treatments: (1)mock-transduction procedure, no vector control (C); (2) non-targeting vector control (NTC); or (3) targeting knockdown targeting knockdown vector (TV). In all groups, cells were exposed to vectors (or an equivalent volume of media for C) at a cell:DRP titer of 1:100,000 in serum-free DMEM containing 2 μM doxorubicin hydrochloride (Calbiochem, La Jolla, CA) for 2-h, followed by addition of 10% FBS24. Preliminary work identified this protocol and cell:DRP titer ratio as the lowest and most efficacious method able to achieve >75% transduction efficiency. After a total of 8 h, media was removed and replaced with supplemented DMEM. Cells were monitored for onset of GFP, which began 48-h post-vector exposure, using fluorescent microscopy and flow cytometry. To evaluate gene expression changes with and without inflammatory insults, cells were exposed to one of three media conditions 24-h prior to harvest: (1) fresh MEM-E, only; (2) LPS in MEM-E, as described; or (3) 10 ng/ml rhIL-1β (Roche, Mannheim, Germany) in MEM-E. All treatment groups were collected from culture flasks 48-h post-onset of GFP [Fig. 2(A)] to allow adequate time for transcript knockdown to occur.
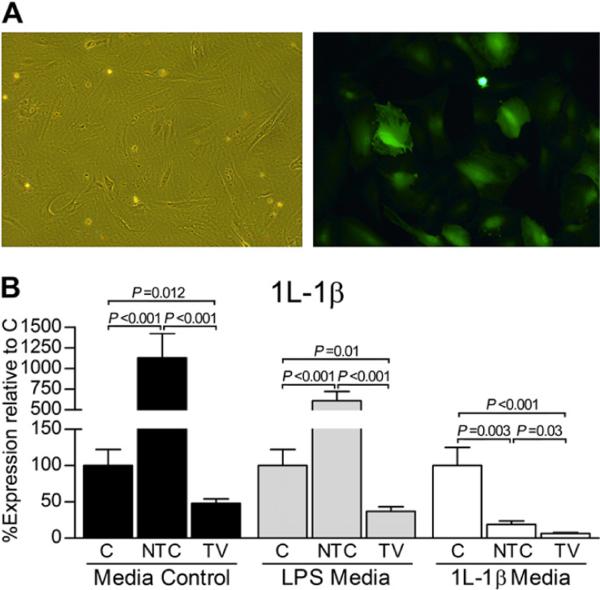
Fig. 2.
(A) Representative photomicrographs of AAV5 vector-transduced guinea pig chondrocytes (200 × final magnification). (B) In vitro AAV5 vector-based reduction of the IL-1β transcript via delivery of the selected shRNA #1 sequence, as indicated in Fig. 1. The targeting knockdown vector (TV) consistently demonstrated significantly reduced IL-1β transcript levels relative to the mock-transduced, no vector control (C) and the non-targeting vector control (NTC). Within each media condition (control, LPS, or IL-1β), fold differences and subsequent percent gene expression levels (mean± 95% confidence intervals) relative to C were calculated using the comparative CT (2−ΔΔCT) method. Data (triplicate assay values for triplicate culture samples) were analyzed using one-way ANOVA for comparison among groups (N, NTC, and TV) followed by pairwise comparisons using Tukey 95% confidence intervals; significant differences are indicated.
In vivo knockdown
Three 2-month-old male Hartley guinea pigs aseptically received TV (1 × 1012 DRP in 100 μl of phosphate buffered saline (PBS)) in one knee, while the opposite knee was injected with an equivalent volume of PBS. An additional three guinea pigs received TV in one knee or an identical vector dose of NTC in the other knee. Administered vector doses were consistent with other peer-reviewed manuscripts25,26. Two months post-injection, animals were humanely euthanized; cartilage was collected from the patella and patellar surface of the femur for RNA extraction to detect IL-1β transcript expression, as described below. One-step reverse transcription polymerase chain reaction (RT-PCR) (Invitrogen, Carlsbad, CA) confirmed GFP expression using specific primers (Table I; Eurofins MWG Operon, Huntsville, AL); glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a positive control. Universal cycling parameters were employed for cDNA construction and amplification. Products were run in a 1% agarose gel containing ethidium bromide and visualized under ultraviolet (UV) light.
Table I.
Guinea pig specific primers used for real-time RT-PCR
| Transcript of interest | Primer sequences (5′–3′) |
|---|---|
| GAPDH* | F: GTATCGTGGAAGGACTCATGACC |
| R: GTTGAAGTCACAGGACACAACCT | |
| eGFP* | F: CATGATATAGACGTTGTGGCTGTTG |
| R: AAGCTGACCCTGAAGTTCATCTGC | |
| IL-1b* | F: ACGCCTGGTGTTGTCTGAC |
| R: GGGAACTGAGCGGATTC | |
| 18S ribosomal RNA† | F: TGCATGGCCGTTCTTAGTTG |
| R: AGTTAGCATGCCAGAGTCTCGTT | |
| IL-2‡ | F: CTTAAGCTCTCCAAAGCA |
| R: CCATCTCTTCAGAAATTCCAC | |
| IL-4§ | F: CATCGGCATTTTGAACGAGGTCA |
| R: CTATCGATGAATCCAGGCATCG | |
| IL-6§ | F: CAGCCCTGAGAAAGGAGACAT |
| R: AATCTGAGGTGCCCATGCTAC | |
| IL-8† | F: GGCAGCCTTCCTGCTCTCT |
| R: CAGCTCCGAGACCAACTTTGT | |
| IL-10‡ | F: GGCACGAACACCCAGTCTGA |
| R: TCACCTGCTCCACTGCCTTG | |
| IL-12‡ | F: TCTGAGCCGGTCACAACTGC |
| R: AGGCGCTGTCCTCCTGACAC | |
| Collagenase 1 (MMP1)∥ | F: AGGTTATCCCAAAATGAT |
| R: TGCAGTTGAACCAGCTATT | |
| Aggrecanase (MMP2)§ | F: AGGGCACCTCCTACAACAGC |
| R: CAGTGGACATAGCGGTCTCG | |
| Collagenase 3 (MMP13)∥ | F: TTCTGGCACATGCTTTTCCTC |
| R: GGTTGGGGTCTTCATCTCCTG | |
| TGF-β† | F: CATCGATATGGAGCTGGTGAAG |
| R: GCCGTAATTTGGACAGGATCTG | |
| TNF-α† | F: CCTACCTGCTTCTCACCCATACC |
| R: TTGATGGCAGAGAGAAGGTTGA | |
| IFN-γ† | F: ATTTCGGTCAATGACGAGCAT |
| R: GTTTCCTCTGGTTCGGTGACA | |
| iNOS (LT)¶ | F: TGGATGCAACCCCATTGTC |
| R: CCCGCTGCCCCAGTTT |
Primers designed by KSS. All amplicons were sequenced to confirm gene transcript specificity.
Allen SS, et al. Infect Immun 2003;71(8):4271-7.
Yamada H, et al. Exp Anim 2005;54(2):163-172.
Oh C, et al. Am J Obstet Gynecol 2008;199(1):78.e1-6.
Huebner JL, et al. Arthritis Rheum 1998;41(5):877-90.
Thompson L, et al. Pediatr Res 2009;65(2):188-92.
Flow cytometry
Chondrocytes were analyzed for and, for plasmid-transfected cells, separated by GFP using the i-Cyt Reflection (i-Cyt, Champaign, IL). Green fluorescence was excited by an argon 488 nm laser and detected using a 520/30 nm bandpass filter on the 520 parameter; positive cells were collected for analysis.
Relative real-time (q)RT-PCR
Total RNA was isolated from in vitro and in vivo chondrocytes via the NucleoSpin® RNA XS kit (Clontech, Mountain View, CA), which includes DNase treatment. cDNA was made using Taqman® Reverse Transcription Reagents (Applied Biosystems, Foster City, CA) and qRT-PCR, complete with dissociation curve, was performed in triplicate for each gene of interest under each triplicate experimental condition (triplicate assay values for triplicate culture samples for in vitro work; triplicate assay values for N = 6 animals) using Power SYBR® Green PCR Master Mix (Applied Biosystems, Warrington, UK) and guinea pig specific primers (Table I) on the ABI Prism® 7000 Sequence Detection System (Applied Biosystems, Foster City, CA) according to published standards and recommendations27. 18s ribosomal RNA and GAPDH were used as separate endogenous controls to which each gene of interest was normalized. Results were consistent between housekeeping genes; GAPDH comparisons are represented. To confirm absence of amplification, reactions were repeated using larger starting amounts of cDNA. Fold changes and subsequent percent gene expression levels relative to designated control groups were calculated using the comparative CT (2−ΔΔCT) method. For plasmid-transfection analyses, potential targeting shRNA plasmids of interest were compared to the non-targeting shRNA control sequence within each media. To stringently evaluate TV, all comparative CT values were normalized relative to C within each media (MEM-E, LPS in MEM-E, or rhIL-1β in MEM-E). In vivo comparative CT values for TV were compared to control knee cartilage receiving saline-only or NTC injection.
Statistical analysis
In vitro (triplicate assay values for triplicate culture samples) and in vivo (triplicate assay values for N = 6 animals) ΔCT values for each gene of interest under each experimental condition were analyzed for – and passed – Gaussian distribution and homogeneous variance using the Kolmogorov–Smirnov normality test28. Flow cytometry and relative percent expression qRT-PCR data (directly converted from fold changes) were expressed as mean ± 95% confidence interval. For in vitro data, relative qRT-PCR data within and between media conditions were analyzed for each individual gene of interest by one-way analysis of variance (ANOVA) for comparison among groups (C, NTC, and TV) followed by pairwise comparisons using Tukey 95% confidence intervals. Paired t-tests were utilized for in vivo comparisons within C/TV and NTC/TV groups; statistical differences in ΔCT values were not detected within or between animals. All analyses were performed using the Minitab statistical software program (State College, PA) with a statistical significance of P < 0.05.
Results
Preliminary studies to detect IL-1β in 2-month-old guinea pig chondrocytes at transcript and protein levels via RT-PCR and immunohistochemistry, respectively, confirmed presence in host tissue and cultured cells up to the fourth passage [Fig. 1(B)]. Flow cytometry revealed that 28.15± 2.44% of plasmid-transfected cells were positive for GFP [Fig. 1(C)], which is consistent with other lipid-based DNA delivery methods for primary chondrocytes29. Relative qRT-PCR identified three of ten shRNA sequences that significantly decreased IL-1β transcript levels relative to the non-targeting control plasmid [Fig. 1(D)]. In control media, shRNA #1 achieved a degree of knockdown (89.42± 0.38%) that was greater (P = 0.004) than the reduction provided by shRNA#2 and #3 and was anticipated to influence expression of other genes of interest. This knockdown (97.18±2.72%) was maintained (P = 0.002) during active LPS challenge [Fig. 1(D)].
The influence of IL-1β knockdown on expression of other mediators was comparable between plasmid-transfected and vector-transduced chondrocytes; expression changes for the latter are presented.
In vitro RNAi
Flow cytometry on pilot studies revealed that 87.50± 4.55% of chondrocytes expressed GFP 48 h following initial 2-h application of AAV5 vectors [Fig. 2(A)]. Amplification was not detected for the following transcripts (Table I): collagenase I/matrix metalloproteinase-1 (MMP1); IL-4; IL-6; and IL-10 (data not shown).
IL-1β transcript
Media influence
Exogenous LPS increased (P < 0.001) transcript expression of IL-1β relative to control media (Table II). Exogenous rhIL-1β, however, decreased (P = 0.007) expression of IL-1β relative to control media (Table II), and treatment with either AAV5 vector significantly reduced this expression further (Fig. 2).
Table II.
ΔCT values ± Δ standard deviation (normalized to GAPDH) and corresponding percent expression (mean ± standard deviation) of transcripts of interest in inflammatory media conditions relative to control media
| Transcript | Control media |
LPS media |
rhIL-1β media |
|||
|---|---|---|---|---|---|---|
| ΔCT ± ΔSD | % ± SD | ΔCT ± ΔSD | % ± SD | ΔCT ± ΔSD | % ± SD | |
| IL-1β | 2.82 ± 0.12 | 100 ± 8.67 | 1.22 ± 0.10 | 303.41 ± 20.26 (P< 0.001) | 3.60 ± 0.14 | 58.24 ± 5.39 (P =0.007) |
| TNF-α | 7.22 ± 0.14 | 100 ± 9.38 | 1.20 ± 0.10 | 6,489.34 ± 434.57 (P< 0.001) | 2.22 ± 0.12 | 3,244.67 ± 263.10 (P< 0.001) |
| IL-2 | ND | – | ND | – | 8.66 ± 0.14 | NA |
| IL-8 | 4.62 ± 0.13 | 100 ± 8.72 | 0.11 ± 0.10 | 2,294.33 ± 168.74 (P< 0.001) | 1.11 ± 0.11 | 1,147.16 ± 84.21 (P< 0.001) |
| IL-12 | 5.10 ± 0.14 | 100 ± 9.63 | ND | – | 4.70 ± 0.14 | 131.19 ± 11.45 (P =0.009) |
| TGF-β | −4.91 ± 0.10 | 100 ± 7.17 | −3.50 ± 0.12 | 37.89 ± 3.03 (P =0.003) | −3.12 ± 0.10 | 28.71 ± 1.92 (P =0.002) |
| MMP13 | 2.94 ± 0.11 | 100 ± 7.92 | −3.19 ± 0.08 | 7,003.48 ± 377.79 (P< 0.001) | −1.4 ± 0.10 | 2,082.15 ± 139.44 (P< 0.001) |
| MMP2 | ND | – | 4.32 ± 0.11 | NA | 6.00 ± 0.10 | NA |
| IFN-γ | −8.12 ± 0.14 | 100 ± 9.25 | −8.71 ± 0.02 | 151.57 ± 2.09 (P =0.005) | −9.60 ± 0.08 | 282.84 ± 15.26 (P< 0.001) |
| iNOS | 3.16 ± 0.14 | 100 ± 9.34 | 2.42 ± 0.08 | 167.01 ± 9.01 (P =0.003) | 2.22 ± 0.11 | 191.85 ± 14.09 (P< 0.001) |
ND =Amplicon not detected; NA= Comparative CT(2−ΔΔCT) method not applicable; P values are indicated.
Transcript changes
Significant decreases in IL-1β expression were achieved by TV relative to C and NTC groups in all media [Fig. 2(B)]. Specifically, IL-1β was reduced to 48.00 ±2.54%, 37.25 ±1.44%, and 6.50 ± 0.29% relative percent expression levels in control, LPS, and rhIL-1β media, respectively. This knockdown is particularly striking given that NTC increased (P < 0.001) transcript levels of IL-1β in control and LPS media. Although exogenous rhIL-1β statistically decreased IL-1β expression (which was further exaggerated in the presence of AAV5 vector), TV continued to result in a decreased IL-1β transcript level relative to C (P < 0.001) and NTC (P = 0.03) control groups.
Inflammatory mediators
Media influence
Overall, exogenous LPS and rhIL-1β significantly increased transcript expression of inflammatory genes of interest relative to control media (Table II). IL-2, however, was only detected in media containing rhIL-1β [Fig. 3(B)] and LPS media reduced expression of IL-12 to below detection limits [Fig. 3(D)].
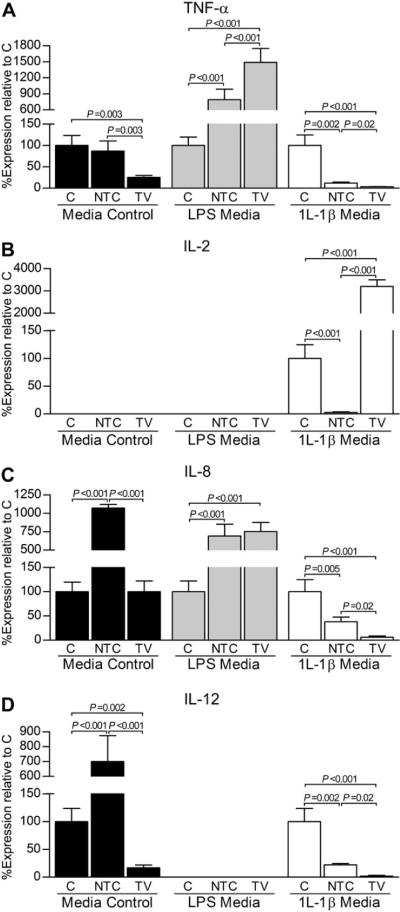
Fig. 3.
Associated transcript expression changes for inflammatory mediators TNF-α (A), IL-2 (B), IL-8 (C), and IL-12 (D) following in vitro AAV5 vector-based reduction of the IL-1β transcript. Within each media condition (control, LPS, or IL-1β), fold differences and subsequent percent gene expression levels (mean ± 95% confidence intervals) for the targeting knockdown vector (TV) and non-targeting vector control (NTC) relative to the mock-transduced, no vector control (C) were calculated using the comparative CT (2−ΔΔCT) method. Data (triplicate assay values for triplicate culture samples) were analyzed using one-way ANOVA for comparison among groups (N, NTC, and TV) followed by pairwise comparisons using Tukey 95% confidence intervals; significant differences are indicated.
Influence of RNAi
Transcript changes associated with inflammatory agents were significantly affected following exposure to TV and NTC, and these changes were often influenced by ambient media that was present. In general, transduction with TV statistically decreased transcript levels of key inflammatory cytokines relative to C and NTC groups. Findings relevant to individual genes and media are elucidated below.
Tumor necrosis factor-α (TNF-α)
In control and rhIL-1β media, TNF-α transcript levels were significantly reduced by TV relative to C and NTC control groups [Fig. 3(A)]. In rhIL-1β media, TNF-α was decreased (P = 0.002) by NTC relative to C, but reduction continued to be most significant by TV. In LPS media, however, TNF-α was increased (P < 0.001) in TV and NTC groups compared to C. Further, TNF-α was greater (P < 0.001) in TV treated cells than NTC-treated cells.
IL-2
IL-2 was detected only in rhIL-1β media treatment groups [Fig. 3(B)]. IL-2 was increased (P < 0.001) by TV compared to NTC and C control groups.
IL-8
Expression patterns observed for the IL-8 transcript were influenced by media conditions [Fig. 3(C)]. In control media, IL-8 levels were increased (P < 0.001) by NTV relative to TV and C. No statistical difference was found between the latter two treatments. In LPS media, however, TV and NTC groups were increased (P < 0.001) relative to C. Exogenous rhIL-1β media, on the other hand, decreased (P < 0.001) the IL-8 transcript in the presence of either AAV5 vector, with a significant reduction (P = 0.02) induced by TV.
IL-12
IL-12 was present in control and rhIL-1β media, only, and was statistically lower in TV relative to C and NTC control groups in both conditions [Fig. 3(D)]. In control media, IL-12 was increased (P < 0.001) in NTC relative to C, while it was decreased (P = 0.002) by NTC relative to C in the presence of rhIL-1β.
Control media
In general, transcript levels of inflammatory cytokines in control media were either below detection or demonstrated a significant decrease in the presence of TV (Fig. 3). Interestingly, IL-8 and IL-12 were significantly increased when transduced by NTC relative to C.
LPS media
In LPS media, IL-2 and IL-12 were below detection limits. TNF-α [Fig. 3(A)] and IL-8 [Fig. 3(C)], however, showed increased (P < 0.001) cytokine transcript expression in the presence of either the TV or NTC AAV5 vector. TNF-α, in particular, was further increased (P < 0.001) in TV treated cells exposed to LPS media relative to NTC cells.
rhIL-1β media
All transcripts of interest, except for IL-2, were statistically decreased by TV in exogenous rhIL-1β media (Fig. 3). For IL-2, transduction with TV resulted in an increase (P < 0.001) in IL-2 transcription relative to C and NTC control groups [Fig. 3(B)].
Anabolic and catabolic agents
Media influence
Overall, exogenous LPS and rhIL-1β significantly decreased transcript expression of transforming growth factor-β (TGF-β) relative to control media (Table II). LPS and rhIL-1β treatment, however, statistically increased transcript numbers of all catabolic agents investigated (Table II). Gelatinase A/MMP2 was only detected in control media following exposure to NTC [Fig. 5(B)].
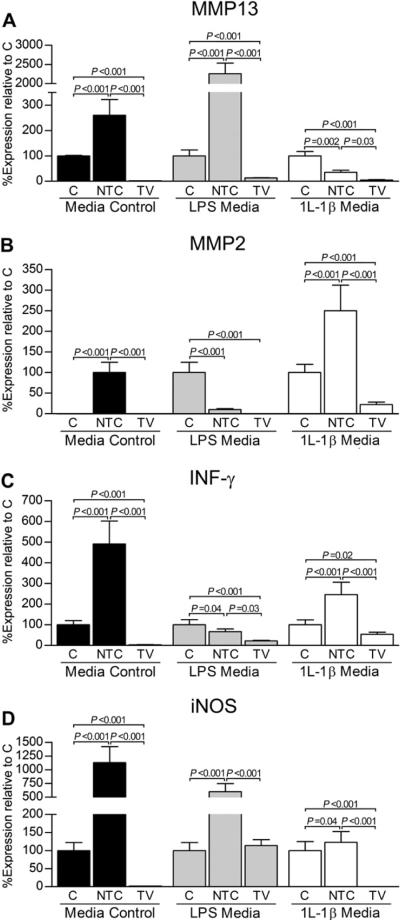
Fig. 5.
In vitro reduction of the IL-1β transcript influences transcript expression of the catabolic mediators MMP13 (A), MMP2 (B), IFN-γ (C), and iNOS (D). In general, transduction with the targeting knockdown vector (TV) significantly reduced the genes of interest in all media conditions relative to the mock-transduced, no vector control (C) and non-targeting vector control (NTC). Within each media condition (control, LPS, or IL-1β), fold differences and subsequent percent gene expression levels (mean ± 95% confidence intervals) for TV and NTC relative to C were calculated using the comparative CT (2−ΔΔCT) method. Data (triplicate assay values for triplicate culture samples) were analyzed using one-way ANOVA for comparison among groups (N, NTC, and TV) followed by pairwise comparisons using Tukey 95% confidence intervals; significant differences are indicated.
Influence of RNAi
In general, transcript levels of anabolic TGF-β were significantly increased by use of TV relative to C and NTC control groups. Transduction with TV also statistically decreased transcript levels of catabolic agents relative to C and NTC. Results pertinent to individual genes and media are discussed below.
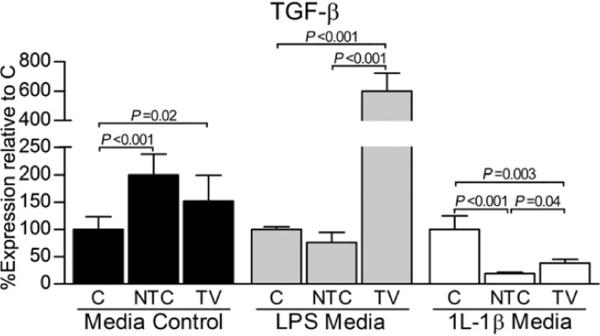
TGF-β
In LPS and rhIL-1β media, expression of TGF-β was significantly increased by TV relative to NTC (Fig. 4). Of note, TV resulted in an increase (P < 0.001) in TGF-β relative to C in LPS media. In control media, both AAV5 vectors statistically increased TFG-β transcript levels relative to C; a significant difference between these two AAV5 vectors was not present.
Fig. 4.
In vitro reduction of the IL-1β transcript influences transcript expression of the anabolic agent TGF-β. Within each media condition (control, LPS, or IL-1β), fold differences and subsequent percent gene expression levels (mean ± 95% confidence intervals) for the targeting knockdown vector (TV) and non-targeting vector control (NTC) relative to the mock-transduced, no vector control (C) were calculated using the comparative CT (2−ΔΔCT) method. Data (triplicate assay values for triplicate culture samples) were analyzed using one-way ANOVA for comparison among groups (N, NTC, and TV) followed by pairwise comparisons using Tukey 95% confidence intervals; significant differences are indicated.
Collagenase III/MMP13
In all media, MMP13 was significantly reduced following transduction with TV [Fig. 5(A)]. In contrast, MMP13 was statistically increased by NTC relative to C in control and LPS media. In rhIL-1β media, MMP13 was decreased in both AAV5 vector treatment groups relative to C, but TV continued to demonstrate fewer (P = 0.03) MMP13 transcripts relative to NTC.
Gelatinase A/MMP2 and interferon-γ (IFN-γ)
Expression patterns for MMP2 and IFN-γ were similar. In both cases, gene expression was significantly decreased by TV relative to C and NTC control groups in LPS and rhIL-1β media [Fig. 5(B and C)]. Further, the IFN-γ transcript was reduced (P < 0.001) by TV relative to C in control media. While NTC statistically decreased transcript levels relative to C in LPS-treated chondrocytes, MMP2 and IFN-γ transcripts were significantly increased by NTC in rhIL-1β media.
Inducible nitrous oxide synthase (iNOS)
In control and rhIL-1β media, iNOS was below detection limits following treatment with TV [Fig. 5(D)]. For LPS media, iNOS was not different between TV and C groups; transcript levels, however, were significantly higher (P < 0.001) in the NTC group. In all media, transduction with NTC resulted in statistically increased expression levels of iNOS relative to TV and C.
Control media
In control media, treatment with either AAV5 vector resulted in a statistical increase in TGF-β transcript numbers (Fig. 4). With the exception of MMP2, for which expression was only detected following NTC treatment, all catabolic genes were significantly reduced by TV relative to C and NTC (Fig. 5). Of note, transcript numbers for all catabolic genes were increased when transduced by NTC relative to C.
LPS media
In LPS media, expression of TGF-β was increased (P < 0.001) by TV relative to C and NTC (Fig. 4). Except for iNOS, transcript levels of all catabolic agents in this media were either below detection or demonstrated statistical decreases following exposure to TV (Fig. 5).
rhIL-1β media
Treatment with either AVV5 vector resulted in significantly reduced TGF-β transcript numbers relative to C (Fig. 4). Importantly, expression of TGF-β was decreased (P = 0.04) by TV relative to NTC in this media. Transcript levels of all catabolic agents following exposure to TV demonstrated statistical reductions relative to C and NTC (Fig. 5). Except for MMP13, remaining catabolic mediators were also significantly increased by NTC relative to C.
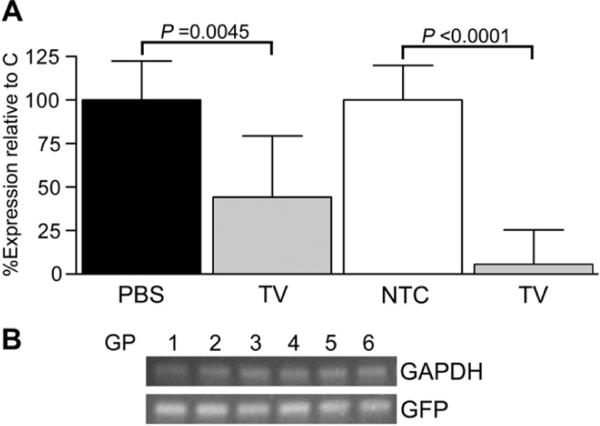
In vivo RNAi
A decrease (P = 0.0045) in IL-1β transcript level (44.25 ± 8.16%) was attained in cartilage in vivo by TV relative to PBS-only, contralateral control knees [Fig. 6(A)]. A reduction (P = 0.0001) in IL-1β (5.50 ± 8.00%) was also present in cartilage exposed to TV relative to contralateral NTC-treated knees. Importantly, reporter gene expression was detected in cartilage harvested from knees injected with TV or NTC [Fig. 6(B)].
Fig. 6.
(A) In vivo transduction in six 2-month-old guinea pigs (GP) with the targeting knockdown (TV) AAV5 vector resulted in statistically significant reductions in IL-1β transcript expression relative to saline-only control (PBS; GP1–3) or non-targeting control vector (NTC; GP4–6) exposed cartilage in opposite knees. Fold differences and subsequent percent gene expression levels (mean ± 95% confidence intervals) for TV relative to PBS or NTC were calculated in the six animals (N = 3 in the group receiving either TV or PBS and N = 3 in the group receiving TV or NTC). For each treatment condition and individual animal, triplicate assay replicates for the gene of interest were determined via real-time RT-PCR utilizing the comparative CT (2−ΔΔCT) method and analyzed using paired t-tests; significant differences are indicated. (B) Expression of GFP was confirmed by RT-PCR in each knee injected with TV (shown) or NTC (data not shown); GAPDH served as a positive internal control.
Discussion
We recently described the temporal expression and tissue distribution of IL-1β through progression of primary OA in two strains of guinea pigs with varying disease propensity30. Persistent IL-1β was found in cartilage, menisci, synovium, and subchondral bone in OA-prone animals at 120 and 180 days, while OA-resistant animals demonstrated a significant decrease in expression, a finding that suggested a relationship between IL-1β and premature disease incidence in the former strain. These findings prompted the current work, which had a primary aim of identifying a tool capable of reducing IL-1β-mediated pathways such that evidence of its mechanistic contribution to OA could be determined.
To our knowledge, this is the first study to demonstrate AAV5 vector-based reduction of the IL-1β transcript via U6-driven shRNA-mediated RNAi in OA-prone guinea pig chondrocytes. Importantly, cells in vitro were exposed to two standard inflammatory agents, LPS and rhIL-1β, to demonstrate persistent knockdown during biologically relevant scenarios where an upregulation of the IL-1β transcript is anticipated. This knockdown also resulted in statistically significant changes in gene expression of other factors implicated in OA pathogenesis in all media conditions. Of note, key inflammatory (TNF-α, IL-2, IL-8, and IL-12) and catabolic agents (MMP13, MMP2, IFN-γ, and iNOS) were significantly decreased by TV relative to C and NTC, while an anabolic factor (TGF-β) was statistically increased. Further, in vivo application of TV revealed that IL-1β levels were reduced >50% relative to contralateral control cartilage and >90% relative to NTC-exposed cartilage. A long-term study, including additional timepoints and an increased sample size, is merited to pursue associated genes of interest that may be influenced by this degree of in situ transcript reduction in cartilage, as well as other relevant joint tissue.
In general, transcript patterns observed following targeted reduction of IL-1β via RNAi in guinea pig chondrocytes coincided with findings exhibited following in vitro treatment with diacerein/rhein or IRAP, particularly in regards to MMPs, iNOS, and TGF-β. For example, diacerein/rhein has been shown to decrease secreted protein levels of proMMPs-1, -3, and -13 in cultured rabbit articular chondrocytes exposed to rhIL-1α at a concentration of 1 ng/ml6. Human OA cartilage treated with exogenous rhIRAP reduces these same mediators at the transcript level7. Further, in equine synoviocytes transduced with an adenoviral vector carrying a coding region for IRAP, MMP13 mRNA expression was significantly lower compared to non-transduced controls8. Interestingly, however, one study evaluating diacerein did not detect a significant difference in MMP13 in media of human OA chondrocytes stimulated by rhIL-1β (10 U/ml), but differences in experimental design may account for this discrepancy9. Reduction of iNOS mRNA and NO release from bovine chondrocytes10, human OA chondrocytes in alginate beads11, and LPS-stimulated (1 mg/ml) human OA cartilage12 have also been observed following application of diacerein/rhein in a dose-dependent manner. The study utilizing human cells in alginate beads also demonstrated that, in media containing 10−10 M rhIL-1β, rhein did not significantly modify IL-8 production11. Enhanced expression of TGF-β1 and TGF-β2 mRNA and total mature TGF-β protein were observed in bovine chondrocytes treated with 10 ng/ml IL-1β (species source not indicated)13, as well as rabbit chondrocytes14. Corroboration of our findings with other work provides evidence that use of vector-based shRNA to reduce the effective concentration of the IL-1β transcript in chondrocytes may be equivalent to established methods.
TNF-α in LPS media and IL-2 in rhIL-1β media were statistically increased by TV relative to C and NTC controls. Although there are no studies that have previously discussed the effects of IL-1β antagonism on IL-2 expression, a study using equine synoviocytes in basal media and media containing 10 ng/ml IL-1β demonstrated that TNF-α was significantly increased following transduction with an adenoviral vector containing a coding region for IRAP, particularly when exposed to the latter treatment15. Additional work is necessary to establish mechanisms leading to increases in these inflammatory cytokines with concurrent IL-1β reduction and in relation to specific media. Our findings emphasize complex and opposing effects that ambient environmental stimulators have on gene expression in the face of identical treatment protocols.
Our study also demonstrated that NTC significantly increased transcript levels of IL-1β, TNF-α, IL-8, IL-12, MMP13, MMP2, IFN-γ, and iNOS in various media relative to C and TV. AAV, itself, is known to be a weak innate immunogen, and cytokine and chemokine responses in transduced tissues are limited and highly transient16. For instance, intravenous administration of an AAV vector resulted in rapid induction of TNF-α mRNA in murine liver cells that returned to normal levels within 6 h17. As such, the significance of inflammatory transcript changes noted in our in vitro study may not have relevant in vivo implications. The influence that a non-targeting control AAV5 vector may have on catabolic agents, however, is unknown and merits consideration.
Published qRT-PCR primer sets for MMP1, IL-4, IL-6, and IL-10 were validated in our laboratory and tested throughout this study, but did not result in detectable amplicons. These findings may, indeed, indicate that these mediators were not undergoing transcription, but further work continues to trouble-shoot assay conditions and assess alternate primer options. We were particularly interested in MMP1, as differing effects related to IL-1β antagonists have been reported. In one study, rhein reduced transcript levels of this catabolic agent in rhIL-1β (10 ng/ml) stimulated bovine chondrocytes18, while another study using rhIL-1β (10 U/ml) treated human OA chondrocytes were exposed to diacerein and a significant reversal in MMP1 levels was not detected in culture media9. In contrast, exogenous hrIRAP has shown a dose-dependent decrease in MMP1 mRNA in the canine model of experimental OA19. Further, IL-6 production in human chondrocytes was statistically decreased by rhein in unconditioned media but was not affected by diacerein in media containing 10−10 M rhIL-1β11.
Media was collected from treatment groups to correlate transcript changes to detectable protein levels. Given need to exhaustively validate reagent cross-reactivity with guinea pig epitopes, as well as account for inherent lability of several mediators31, studies persist to correlate mRNA data with protein data via Western blot and/or enzyme-linked immunosorbent assay (ELISA) analysis. To date, our laboratory has tested several commercially available ELISA kits designed for use with human, mouse, and rat epitopes, but guinea pig IL-1β levels continue to be below the detection limit of these assays.
In conclusion, a plasmid- and AAV5 vector-based shRNA sequence was capable of reducing the IL-1β transcript in OA-prone chondrocytes in vitro with and without active inflammatory challenges. Differential gene expression of known disease mediators occurred as a result of knockdown, with influences in the direction of beneficial modification of OA pathogenesis. Importantly, in vivo application of TV resulted in a significant reduction of the IL-1β transcript relative to PBS-only or NTC-exposed cartilage. Investigations to characterize expression changes influenced by reduction of the IL-1β transcript in vivo are warranted.
Acknowledgments
K.S. Santangelo was funded by a NIH NIAMS F32 (AR053805) throughout the majority of the work and is currently supported by a fellowship for residency training from GlaxoSmithKline organized by the American College of Veterinary Pathologists and Society of Toxicologic Pathology Coalition for Veterinary Pathology Fellows. The Comparative Orthopedics Laboratory also receives support through the Trueman Family Endowment. The original plasmids used in this study were kindly provided by the Viral Vector Core at Nationwide Children's Research Hospital. We thank Jeffrey Bartlett PhD, Gerard Nuovo MD, and Stephen Weisbrode VMD, PhD for their comments throughout this project. We would also like to acknowledge Spencer Smith, Sarah Baker, Nicole White, and Marc Hardman for technical assistance.
Footnotes
Contributions Kelly S. Santangelo: design of the study, obtaining of funding, acquisition of the data, analysis and interpretation, drafting and revising of the article, and final approval. Responsibility for integrity of the work.
Alicia L. Bertone: design of the study, obtaining of funding, analysis and interpretation, revising of the article, and final approval. Responsibility for integrity of the work.
Conflict of interest The authors declare that there is no conflict of interest.
References
- 1.Clements KM, Price JS, Chamber MG, Visco DM, Poole AR, Mason RM. Gene deletion of either interleukin-1beta, interleukin-1beta-converting enzyme, inducible nitric oxide synthase, or stromelysin 1 accelerates the development of knee osteoarthritis in mice after surgical transection of the medial collateral ligament and partial medial meniscectomy. Arthritis Rheum. 2003 Dec;48(12):3452–63. doi: 10.1002/art.11355. [DOI] [PubMed] [Google Scholar]
- 2.Evans CH, Gouze JN, Gouze E, Robbins PD, Ghivizzani SC. Osteoarthritis gene therapy. Gene Ther. 2004 Feb;11(4):379–89. doi: 10.1038/sj.gt.3302196. [DOI] [PubMed] [Google Scholar]
- 3.Goldring SR, Goldring MB. The role of cytokines in cartilage matrix degeneration in osteoarthritis. Clin Orthop Relat Res. 2004 Oct;(427 Suppl):S27–36. doi: 10.1097/01.blo.0000144854.66565.8f. [DOI] [PubMed] [Google Scholar]
- 4.Caron JP, Fernandes JC, Martel-Pelletier J, Tardif G, Mineau F, Geng C, et al. Chondroprotective effect of intra-articular injections of interleukin-1 receptor antagonist in experimental osteoarthritis. Suppression of collagenase-1 expression. Arthritis Rheum. 1996;39(9):1535–44. doi: 10.1002/art.1780390914. [DOI] [PubMed] [Google Scholar]
- 5.Trippel SB, Ghivizzani SC, Nixon AJ. Gene-based approaches for the repair of articular cartilage. Gene Ther. 2004 Feb;11(4):351–9. doi: 10.1038/sj.gt.3302201. [DOI] [PubMed] [Google Scholar]
- 6.Tamura T, Kosaka N, Ishiwa J, Sato T, Nagase H, Ito A. Rhein, an active metabolite of diacerein, down-regulates the production of pro-matrix metalloproteinases-1, -3, -9 and -13 and up-regulates the production of tissue inhibitor of metalloproteinase-1 in cultured rabbit articular chondrocytes. Osteoarthritis Cartilage. 2001 Apr;9(3):257–63. doi: 10.1053/joca.2000.0383. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi M, Squires GR, Mousa A, Tanzer M, Zukor DJ, Antoniou J, et al. Role of interleukin-1 and tumor necrosis factor alpha in matrix degradation of human osteoarthritic cartilage. Arthritis Rheum. 2005 Jan;52(1):128–35. doi: 10.1002/art.20776. [DOI] [PubMed] [Google Scholar]
- 8.Nixon AJ, Haupt JL, Frisbie DD, Morisset SS, McIlwraith CW, Robbins PD, et al. Gene-mediated restoration of cartilage matrix by combination insulin-like growth factor-I/interleukin-1 receptor antagonist therapy. Gene Ther. 2005 Jan;12(2):177–86. doi: 10.1038/sj.gt.3302396. [DOI] [PubMed] [Google Scholar]
- 9.Alvarez-Soria MA, Herrero-Beaumont G, Sánchez-Pernaute O, Bellido M, Largo R. Diacerein has a weak effect on the catabolic pathway of human osteoarthritis synovial fibroblast – comparison to its effects on osteoarthritic chondrocytes. Rheumatology (Oxford) 2008 May;47(5):627–33. doi: 10.1093/rheumatology/ken116. [DOI] [PubMed] [Google Scholar]
- 10.Mendes AF, Caramona MM, de Carvalho AP, Lopes MC. Diacerhein and rhein prevent interleukin-1beta-induced nuclear factor-kappaB activation by inhibiting the degradation of inhibitor kappaB-alpha. Pharmacol Toxicol. 2002 Jul;91(1):22–8. doi: 10.1034/j.1600-0773.2002.910104.x. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez C, Mathy-Hartert M, Deberg MA, Ficheux H, Reginster JY, Henrotin YE. Effects of rhein on human articular chondrocytes in alginate beads. Biochem Pharmacol. 2003 Feb 1;65(3):377–88. doi: 10.1016/s0006-2952(02)01485-5. [DOI] [PubMed] [Google Scholar]
- 12.Yaron M, Shirazi I, Yaron I. Anti-interleukin-1 effects of diacerein and rhein in human osteoarthritic synovial tissue and cartilage cultures. Osteoarthritis Cartilage. 1999 May;7(3):272–80. doi: 10.1053/joca.1998.0201. [DOI] [PubMed] [Google Scholar]
- 13.Felisaz N, Boumediene K, Ghayor C, Herrouin JF, Bogdanowicz P, Galerra P, et al. Stimulating effect of diacerein on TGF-beta1 and beta2 expression in articular chondrocytes cultured with and without interleukin-1. Osteoarthritis Cartilage. 1999 May;7(3):255–64. doi: 10.1053/joca.1998.0199. [DOI] [PubMed] [Google Scholar]
- 14.Pujol JP, Felisaz N, Boumediene K, Ghayor C, Herrouin JF, Bogdanowicz P, et al. Effects of diacerein on biosynthesis activities of chondrocytes in culture. Biorheology. 2000;37(1–2):177–84. [PubMed] [Google Scholar]
- 15.Haupt JL, Frisbie DD, McIlwraith CW, Robbins PD, Ghivizzani S, Evans CH, et al. Dual transduction of insulin-like growth factor-I and interleukin-1 receptor antagonist protein controls cartilage degradation in an osteoarthritic culture model. J Orthop Res. 2005 Jan;23(1):118–26. doi: 10.1016/j.orthres.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 16.Nayak S, Herzog RW. Progress and prospects: immune responses to viral vectors. Gene Ther. 2010 Mar;17(3):295–304. doi: 10.1038/gt.2009.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaiss AK, Liu Q, Bowen GP, Wong NC, Bartlett JS, Muruve DA. Differential activation of innate immune responses by adeno-virus and adeno-associated virus vectors. J Virol. 2002 May;76(9):4580–90. doi: 10.1128/JVI.76.9.4580-4590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Domagala F, Martin G, Bogdanowicz P, Ficheux H, Pujol JP. Inhibition of interleukin-1beta-induced activation of MEK/ERK pathway and DNA binding of NF-kappaB and AP-1: potential mechanism for Diacerein effects in osteoarthritis. Biorheology. 2006;43(3–4):577–87. [PubMed] [Google Scholar]
- 19.Ristevski S. Making better transgenic models: conditional, temporal, and spatial approaches. Mol Biotechnol. 2005 Feb;29(2):153–63. doi: 10.1385/MB:29:2:153. [DOI] [PubMed] [Google Scholar]
- 20.Babcock AM, Standing D, Bullshields K, Schwartz E, Paden CM, Poulsen DJ. In vivo inhibition of hippocampal Ca2+/calmodulin-dependent protein kinase II by RNA interference. Mol Ther. 2005 Jun;11(6):899–905. doi: 10.1016/j.ymthe.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 21.Arai Y, Kubo T, Fushiki S, Mazda O, Nakai H, Iwaki Y, et al. Gene delivery to human chondrocytes by an adeno-associated virus vector. J Rheumatol. 2000 Apr;27(4):979–82. [PubMed] [Google Scholar]
- 22.Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol. 1998;72:2224–32. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi S, Mercer SA, Dilley R, Trippel SB. Production of recombinant AAV vectors encoding insulin-like growth factor I is enhanced by interaction among AAV rep regulatory sequences. Virol J. 2009 Jan 7;6:3. doi: 10.1186/1743-422X-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan Z, Zak R, Zhang Y, Ding W, Godwin S, Munson K, et al. Distinct classes of proteasome-modulating agents cooperatively augment recombinant adeno-associated virus type 2 and type 5-mediated transduction from the apical surfaces of human airway epithelia. J Virol. 2004 Mar;78(6):2863–74. doi: 10.1128/JVI.78.6.2863-2874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haas CS, Amin MA, Ruth JH, Allen BL, Ahmed S, Pakozdi A, et al. In vivo inhibition of angiogenesis by interleukin-13 gene therapy in a rat model of rheumatoid arthritis. Arthritis Rheum. 2007 Aug;56(8):2535–48. doi: 10.1002/art.22823. [DOI] [PubMed] [Google Scholar]
- 26.Santangelo KS, Baker SA, Nuovo G, Dyce J, Bartlett JS, Bertone AL. Detectable reporter gene expression following transduction of adenovirus and adeno-associated virus sero-type 2 vectors within full-thickness osteoarthritic and unaffected canine cartilage in vitro and unaffected guinea pig cartilage in vivo. J Orthop Res. 2010 Feb;28(2):149–55. doi: 10.1002/jor.20975. [DOI] [PubMed] [Google Scholar]
- 27.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3(6):1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 28.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006 Feb 22;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stöve J, Fiedler J, Huch K, Günther KP, Puhl W, Brenner R. Lipofection of rabbit chondrocytes and long lasting expression of a lacZ reporter system in alginate beads. Osteoarthritis Cartilage. 2002 Mar;10(3):212–7. doi: 10.1053/joca.2001.0495. [DOI] [PubMed] [Google Scholar]
- 30.Santangelo KS, Pieczarka EM, Nuovo GJ, Weisbrode SE, Bertone AL. Temporal expression and tissue distribution of interleukin-1β in two strains of guinea pigs with varying propensity for spontaneous knee osteoarthritis. Osteoarthritis Cartilage. 2011 Apr;19(4):439–48. doi: 10.1016/j.joca.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huebner JL, Kraus VB. Assessment of the utility of biomarkers of osteoarthritis in the guinea pig. Osteoarthritis Cartilage. 2006;14(9):923–30. doi: 10.1016/j.joca.2006.03.007. [DOI] [PubMed] [Google Scholar]