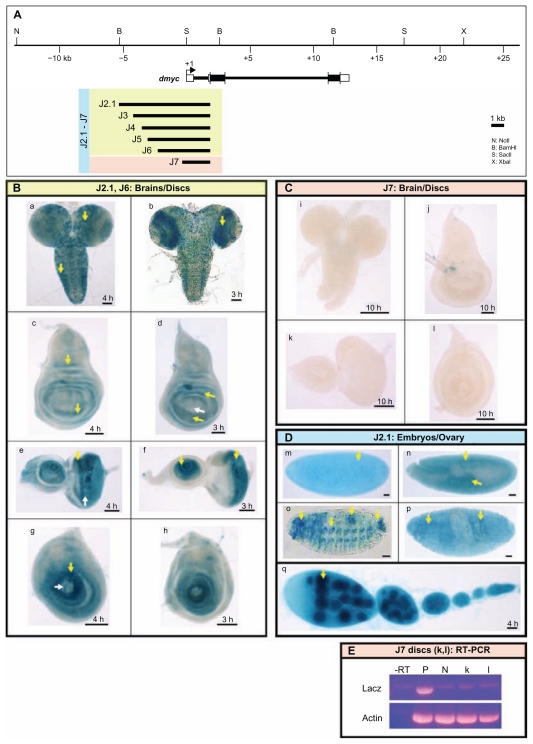
Figure 5.
dmyc 5′ regulatory region contains separable elements. (A) The 40-kb genomic locus and the location of the dmyc is shown at the top. A fragment of 7.159-kb in size (J2.1) (lacking ORF sequences) from the dmyc 5′ region and its deletion restriction fragments were tested for their ability to drive reporter activity in transgenic flies. Colors of the deletion fragments refer to their expression patterns shown in panels B, C and D. (B) J2.1 transgene (a, c, e, g) and all of its successive deletions up to 2.523-kb in the J6 construct (b, d, f, h) were able to express the reporter in a dmyc manner in the brain and discs (a, b: brain; c, d: wing discs; e, f: eye discs; g, h: leg discs). (C) Further truncation down to 1.923-kb of J7 resulted in a loss of expression in the brain and discs (i–l). (D) Shown are embryos and ovary taken from transgenic animals carrying J2.1 transgene. J2.1 retained the expression in the embryos (m–p; embryo stages: m: 2–5; n: 11–12; o, p: 13–16) and ovary (q). Except for 4D o, the smallest construct J7 recapitulated the dmyc like expression in all the other embryonic stages and in ovary. (E) RT-PCR on J7 discs (k, l shown in C), detects no lacZ transcripts beyond background level. Controls used are as follows: –RT: negative control with no transcriptase (brains from Fig. 5B, b); P: Fig. 5B, b, positive control; N: y[1] w[1118] leg discs, negative control; Actin: Drosophila actin as internal control.
Note: Yellow arrow indicates lacZ expression and white arrow indicates lack of lacZ activity. Staining time for discs and ovaries is indicated, and embryos were stained over-night. Scale bar in (a–q) indicates 50 μm.