Abstract
Case-control studies are particularly susceptible to differential exposure misclassification when exposure status is determined following incident case status. Probabilistic bias analysis methods have been developed as ways to adjust standard effect estimates based on the sensitivity and specificity of exposure misclassification. The iterative sampling method advocated in probabilistic bias analysis bears a distinct resemblance to a Bayesian adjustment; however, it is not identical. Furthermore, without a formal theoretical framework (Bayesian or frequentist), the results of a probabilistic bias analysis remain somewhat difficult to interpret. We describe, both theoretically and empirically, the extent to which probabilistic bias analysis can be viewed as approximately Bayesian. While the differences between probabilistic bias analysis and Bayesian approaches to misclassification can be substantial, these situations often involve unrealistic prior specifications and are relatively easy to detect. Outside of these special cases, probabilistic bias analysis and Bayesian approaches to exposure misclassification in case-control studies appear to perform equally well.
Observational research is susceptible to many types of bias. Special attention is given to misclassification, uncontrolled confounding and selection bias, though many other threats to validity exist as well.1,2 Adjusting for biases due to these mechanisms has a long history. Many of the methods specify “bias parameters” (e.g., sensitivity and specificity of misclassification), which dictate the assumed extent of the bias. Historically, authors have attempted to determine how large the bias parameter would have to be in order to make the main effect null.3 Others have specified bias parameters and determined bounds within which the adjusted effect estimate must reside.4,5 More recently, bias parameters have been employed to find bias-adjusted effect estimates. For example, Greenland and Lash6 give formulae to adjust effect estimates for bias due to uncontrolled confounding, misclassification or selection bias.
Recently, many authors have proposed probabilistic bias analysis methods that treat the bias parameters as random variables with probability distributions.6–13 These methods serve to adjust the main effect estimate and propagate uncertainty surrounding the bias parameter, incorporating this uncertainty into the variance estimate of the adjusted main effect. The approach taken in probabilistic bias analysis is to repeatedly draw a random sample from the bias parameter distribution(s) and use those sampled parameters to adjust the effect estimate. The resulting distribution can be summarized with a “bias-adjusted” main effect, as well as uncertainty or simulation intervals.6,8–11,14
Interpretation of the results of a probabilistic bias analysis raises the question of whether they should be viewed under the common frequentist framework or, alternatively, as Bayesian. The explicit use of a distribution for an unknown bias parameter argues strongly for a Bayesian interpretation; indeed, Lash et al 9 interpret probabilistic bias analysis as a semi-Bayes approach in which prior distributions are used for some parameters and not for others. Other authors have implemented simplifications under which a Bayesian interpretation is possible, or performed an outright Bayesian analysis.8,12,13,15–18 However, explicitly Bayesian approaches to bias modeling are difficult to implement (with some exceptions 18,19) and a clear advantage of probabilistic bias analysis over a Bayesian analysis is ease of use. Unfortunately, the extent to which probabilistic bias analysis results mimic Bayesian results remains unclear, with only one published empirical comparison and some theoretical arguments.8,19 In the present paper, we compare probabilistic bias analysis to Bayesian bias modeling for the special case of a dichotomous exposure measured with error in a case-control study.
Methods
We consider a case-control study in which interest focuses on estimating the association between exposure, E, and case/control status, C. We assume y0 of n0 controls (C=0) are classified as exposed while y1 of n1 cases (C=1) are classified as exposed. The true exposure E is misclassified and we observe the apparent exposure, E*. We allow the possibility of differential misclassification and define the sensitivity and specificity of exposure measurement as Pi = Pr(E*=1 | E = 1, C = i) and qi = Pr(E* = 0 | E = 0, C= i), respectively. We further define the measure of etiologic interest, the true odds ratio (OR), as where ri = Pr(E=1 | C = i). An analyst can choose to ignore misclassification of exposure status, and estimate where θi = Pr(E*=1 | C = i). Unfortunately, as is well known, the OR based on apparent exposure status will not generally equal the OR based on true exposure status. If one is willing to assume known fixed values for pi and qi, a simple adjustment is available to calculate a bias adjusted OR:
| (1) |
However, as shown by Gustafson et al,13 an arbitrarily small discrepancy between the true and assumed sensitivities and specificities can lead to an arbitrarily large bias in the adjustment. This is particularly worrying, because it implies that a good guess at sensitivity and specificity may not be sufficient; a perfect guess may be required.
Such concerns, as well as attempts to appropriately incorporate uncertainty in the bias parameter into standard error estimates for the adjusted OR, have lead many authors to treat the sensitivities and specificities as random variables with probability distributions that (ideally) are specified using prior research.6,8,9,11,14 Many choices are available for the distribution of sensitivity and specificity. For the time being, we will remain generic in our specification of the prior parameter distribution and say the sensitivities and specificities have some probability distribution functions, fpi(pi) and fqi(qi), where a choice for f might be the logistic-normal, beta, triangular or trapezoidal distribution. We initially assume the sensitivities and specificities among cases and controls are not correlated for ease of presentation, but relax this assumption in subsequent sections.
Probabilistic bias analysis draws random samples in two steps. The first involves sampling sensitivity and specificity values (p1,p0,q1,q0) from their bias parameter distributions. This is viewed as incorporating uncertainty regarding the extent of the misclassification. Sampled values are plugged into expression (1), replacing θ0 θ1 with the observed exposure probabilities among controls and cases (y0/N0 and y1/N1, respectively), producing an adjusted OR. However, not all combinations of sensitivity and specificity are admissible given the observed data. For instance, if q0 ≤1−θ0 then the adjusted OR could be undefined or negative. Other combinations of sensitivity and specificity are capable of producing similarly nonsensical results. We give bounds for admissible values (p1,p0,q1,q0) in the Appendix. Lash and colleagues9 recommend discarding samples that produce negative cell counts (i.e., samples that lie outside the bounds of admissibility) and drawing new values. Because of these inadmissibility conditions, the distribution of (p1,p0,q1,q0) is dependent on the observed exposure probabilities and we write it as f1 (p1, p0, q1, q0 | θ̂0, θ̂1).
The second step in probabilistic-bias-analysis adjustment for misclassification is to incorporate random sampling error. Standard asymptotic approximations are used to assume that the log of the bias-adjusted OR from the first step has a normally distributed sampling distribution: f2 (log(ψ ) |log(ψc)) ~ N(log(ψc), V(log(ψc))). The variance term is typically estimated using the standard Woolf formula in the unadjusted data, although other options exist.15,20 For each iteration of a probabilistic bias analysis, a random sample is drawn from a normal distribution with a mean equal to the adjusted log-OR obtained in the first step of probabilistic bias analysis. These two steps are iterated a large number of times; resultant adjusted ORs are saved at each iteration and inferences are based on the distribution of the adjusted OR.
A probabilistic-bias-analysis approach bears a distinct resemblance to a Bayesian analysis. The distribution of bias parameters (p1,p0,q1,q0) would be referred to as a prior distribution in a Bayesian analysis. A Bayesian analysis would also specify prior distributions for the parameters r0 and r1. Although in many cases we would suggest specifying informative priors, in this paper we use uniform priors on r0 and r1 to aid in our comparison with probabilistic bias analysis, which typically does not use informative priors. We note that an improper “beta(0,0)” prior with an effective sample size of zero on these parameters could be considered more compatible with the probabilistic-bias-analysis approach. However, it can be verified that this produces an improper posterior in the present context. Thus we instead use the weak uniform prior, i.e. beta(1,1), with an effective sample size of two. Inference from a Bayesian analysis is based on the posterior distribution, which is typically not available in closed-form although samples from it can be generated using Markov chain Monte Carlo (MCMC) algorithm. In the first step, the algorithm draws random samples of sensitivities and specificities from their conditional posterior distribution: g1 ( p1, p0, q1, q0 | θ0, θ1). The samples from step one are then used to generate samples from g2 (θ1, θ0 | p1, p0, q1, q0, y0, y1) in a second step. Neither distribution has a standard form, and Metropolis-Hastings steps can be used to generate these samples. The samples from the two steps are plugged into expression (1) to generate an adjusted OR. The θ1, θ0 from the second step are used in the distribution g1 ( p1, p0, q1, q0 | θ0, θ1 ) at the next iteration. As in probabilistic bias analysis, adjusted ORs from a large number of iterations are collected, and inferences are based on the distribution of those iterates.
The similarity between probabilistic bias analysis and Bayesian approaches to misclassification is intriguing. From a Bayesian perspective, probabilistic bias analysis places a prior distribution on the bias parameters. Both approaches sample iteratively from conditional posterior distributions; i.e., prior distributions that depend on the observed data. The first step in both algorithms samples sensitivities and specificities from the conditional posterior distribution. The second step of both algorithms samples an adjusted OR (either explicitly as in probabilistic bias analysis, or by sampling θ1, θ0 and plugging them into expression (1) as in the Bayesian approach). Finally, inference rests on the distribution of the adjusted OR, referred to as a posterior distribution in the Bayesian approach or a “bias-adjusted” distribution in a probabilistic bias analysis. In the event that the two approaches sample from the same distributions in the first and second steps, we would expect them to give identical results.8
We first focus on the conditional posterior distributions used to sample sensitivities and specificities in the two approaches. To aid in our exposition, we examine only the conditional distribution of specificity in both algorithms (similar results are obtained for the sensitivity parameters). The conditional distribution of specificity in probabilistic bias analysis can be written as f (qi | pi, ci, di, θ̂i) = fqi (qi)IB (qi | θ̑i ), where IB (qi | θ̑i) is an indicator function that takes the value of 1 if the random variable qi falls within the admissibility bounds given in the Appendix (thus ensuring a positive adjusted OR), evaluated at θ̑i = yi / ni. The conditional distribution in probabilistic bias analysis is simply the bias parameter distribution, fqi (qi), truncated to the region implied by the admissibility bounds. If nearly all of the density of the prior distribution lies within these bounds, the truncation will be trivial and the prior and conditional posterior distributions will be nearly identical.
The conditional posterior distribution in a Bayesian analysis is somewhat different: g(qi | pi, ci, di, θi) = fqi (qi) / | pi + qi −1 | IB (qi | θi) There are two important distinctions between this distribution and the one used in a probabilistic bias analysis. First, the distribution in probabilistic bias analysis has a “hard bound” on what values of qi are possible, because the bound depends on θ̂i, which does not vary from iteration to iteration in a probabilistic bias analysis. In contrast, the distribution in the Bayesian analysis has a “soft bound” on qi because values of θi will change at each iteration of the algorithm. Therefore, specificity values that may be impossible to attain in probabilistic bias analysis are possible in the Bayesian analysis.
The second distinction is the division by the scaling factor |pi+qi−1| in the Bayesian approach. The effect of this scaling factor can range from substantial to relatively insignificant. We illustrate the impact of the scaling factor on the conditional posterior distribution of the specificity in Figure 1, where we assume logit (qi) ~ Normal(b, σ2). We show the shape of the prior distribution as well as the shape of the conditional posterior distribution under the probabilistic bias analysis and Bayesian approaches. We illustrate first with a somewhat imprecise prior: b=logit(0.7) and σ2=1; and second with a more precise prior: b=logit(0.7) and σ2=0.05. When pi+qi is close to 1, then the conditional posterior distribution in the probabilistic bias analysis and Bayesian approaches can look considerably different, suggesting the Bayesian and probabilistic bias analysis approaches could produce quite different adjusted ORs. On the other hand, if pi+qi is substantially larger than 1 or prior knowledge is sufficient to produce a relatively compact density function, the scaling factor will have little effect. Intuitively, this rescaling of the prior distribution amounts to redistributing that part of the prior distribution that falls in an inadmissible region (once the data are observed). The inadmissible part of the prior distribution corresponds to a belief in a small (admissible) value of qi. For this reason, prior distributions that fall outside the admissibility bounds will tend to result in conditional posterior distributions with higher density near the bound.
Figure 1.
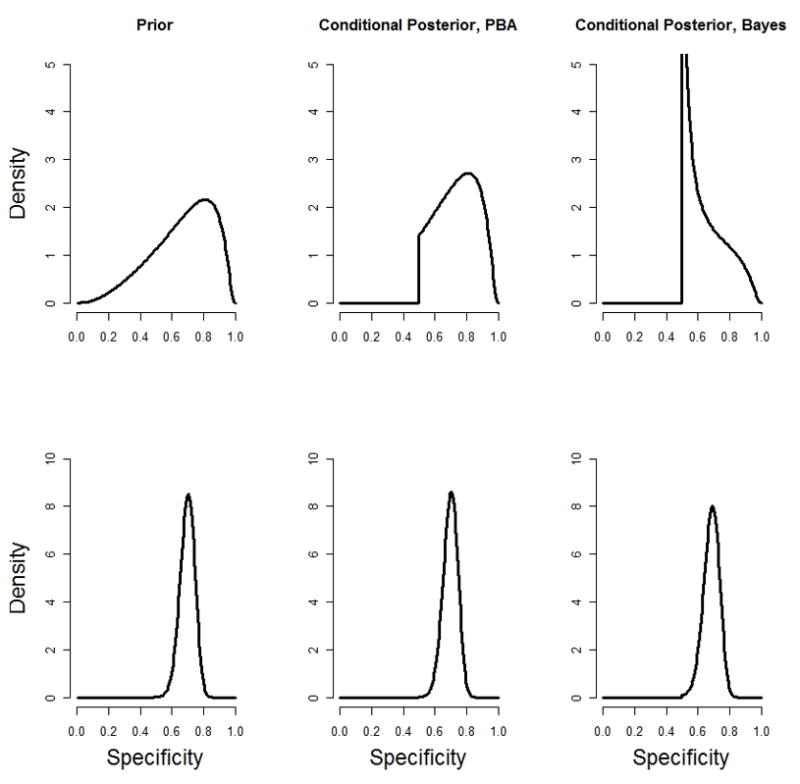
Prior, conditional posterior probabilistic bias analysis (PBA) and conditional posterior Bayesian distributions for specificity parameter. The top row assumes logit(qi) ~ N (logit (0.7),1.0) while the bottom row assumes logit (qi) ~ N (logit(0.7),0.05). All analyses assume pi=0.51 and θi=0.5
The second step of both approaches incorporates random error into the adjusted odds ratio. Probabilistic bias analysis typically relies on an asymptotic argument about the normality of the log-OR, and directly draws samples from this asymptotic distribution. Lash et al9 utilize the Woolf estimator for the variance of the unadjusted log-OR. In contrast, the Bayesian approach updates the observed exposure probabilities (θ0, θ1) and then computes an adjusted OR, using expression (1). Comparison of the two approaches suggests that we could improve the comparability of the two approaches by bootstrapping θ0, θ1 in the second step of probabilistic bias analysis.6,9 The bootstrap estimates would be used to calculate an adjusted OR and would also provide a “soft bound” on the sensitivities and specificities, similar to the Bayesian approach.
Applied Example
We illustrate the potential for differences between the probabilistic bias analysis and Bayesian approach using data from the National Birth Defects Prevention Study, a population-based case-control study of congenital defects. Our interest was in estimating the association between maternal smoking in the periconceptional period (1 month before becoming pregnant to 3 months after) and an infant having an oral facial cleft.21 Exposure misclassification is frequently a concern in case-control studies of birth defects when exposure information is ascertained after the mother knows that her infant has a birth defect. MacLehose et al16 previously reported on a Bayesian approach to exposure misclassification in this study. Here, we compare the results of an explicit Bayesian analysis with a probabilistic-bias-analysis analysis.
Table 1 shows the crude bivariate table of self-reported smoking and case/control status. Unadjusted for any potential misclassification, the data indicate a moderate increased risk of oral facial cleft among infants whose mothers smoked in the periconceptional period (OR=1.4 [95%CI= 1.2–1.7]). A substantial amount of research has been done to evaluate the sensitivity and specificity of self-reported smoking among control mothers.22–24 Unfortunately, we are aware of no previous studies that evaluate the accuracy of self report among mothers of infants with oral facial cleft. In specifying our prior distributions, it is reasonable to expect correlation between sensitivity among cases and controls and correlation between specificity among cases and controls. We follow Chu et al15 in specifying a correlated logistic-normal prior for sensitivities and specificities:
| (2) |
and
Table 1.
Self reported maternal smoking in the periconceptional period and oral facial cleft in the National Birth Defects Prevention Study, 1997–2003
| Reported smoking | Cases (n=1315) | Controls (n=4848) |
|---|---|---|
| Yes | 340 | 949 |
| No | 975 | 3899 |
| (3) |
We complete the specification for the specificity parameters by assuming b0=logit(0.94), b1=logit(0.94), , and ρq = 0.8. These hyperparameter values imply that our best guess at the prior specificity is 0.94 and we are 95% certain the specificity is between 0.92 and 0.95. A correlation of 0.8 has previously been used by Fox et al.14 We choose to apply probabilistic bias analysis and Bayesian approaches to misclassification under 2 sets of priors for sensitivity. Both sets of priors assume a0=logit(0.91), , and ρp = 0.8. This implies that our best guess for the sensitivity among controls is 0.91 and we are 95% certain the sensitivity among controls is between 0.88 and 0.93. However, for the first set of priors we choose a very diffuse prior on the sensitivity among cases: a1=logit(0.5), . For cases, this prior implies our best guess for sensitivity is 0.5 and we are 95% certain the sensitivity is between 0.05 and 0.95. In practice such a vague prior will rarely be realistic and is included here to highlight the potential for divergence between probabilistic bias analysis and Bayesian analysis. Our second prior for sensitivity is altered such that a1=logit(0.91), . This prior implies that our best guess for the sensitivity among cases is 0.91 and we are 95% certain of values between 0.50 and 0.99. This represents a very wide but more defensible prior. A paper by MacLehose et al16 provides more detail on prior specification and alternative prior specifications that address the low specificity of self-reported smoking in this problem.
The Bayesian approach is implemented through a MCMC algorithm similar to the one used by Gustafson and colleagues.13 We ran the algorithm for 10,000 iterations and excluded the initial 1,000 iterations as a burn-in period. We implemented the probabilistic bias analysis as outlined above, with 10,000 iterations of the algorithm (no burn-in period is required with probabilistic bias analysis).
Figure 2 shows the posterior distribution for p1 and log(OR) from the two approaches using the vague prior and weakly informative priors. The vague prior places considerable probability outside the admissibility bounds and, as a result, the posterior distribution of sensitivity in the Bayesian approach favors smaller values than in the probabilistic bias analysis. The data contain information that informs our estimate of sensitivity, and so the Bayesian posterior distribution does not resemble the probabilistic-bias-analysis posterior distribution. The posterior distribution of the log(OR) is notably more peaked for the probabilistic bias analysis, and has a far heavier right tail in the Bayes approach. This is not surprising, since the lower sensitivity values favored in the Bayes approach correspond to higher adjusted ORs. The posterior distributions under the weakly informative prior are also shown in Figure 2. Very little prior density falls outside the admissibility bounds in this example. As a result, the posterior distribution of sensitivity in the Bayes and probabilistic bias analysis approach are virtually identical, as are the posterior log(OR) distributions.
Figure 2.
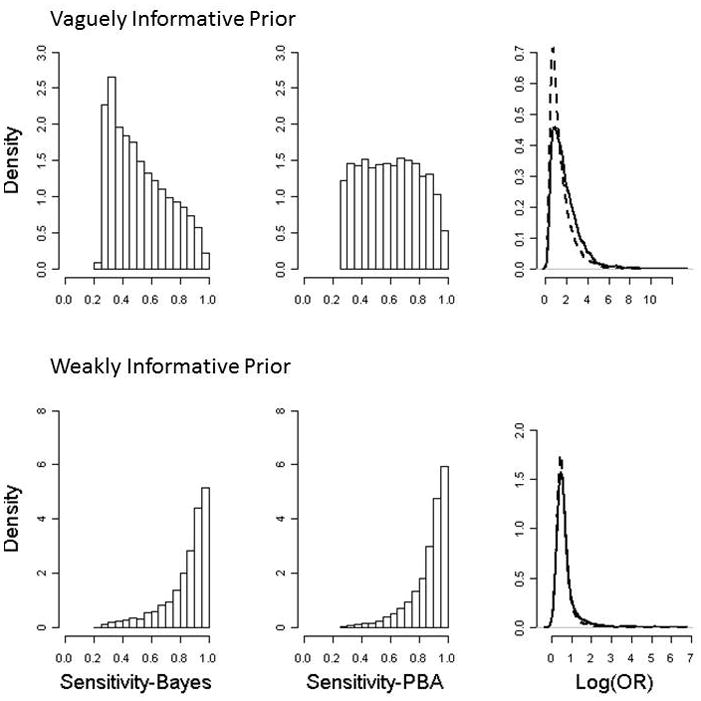
Posterior distribution of sensitivity and log(OR) under the Bayesian and probabilistic bias analysis approaches. First row results from a vaguely informative prior on sensitivity while the second row results from a weakly informative prior. Dashed lines are the density estimates for the probabilistic bias analysis approach and solid lines are for the Bayesian approach.
Simulation Study
Although analysis of a single dataset is instructive, we also examine the performance of the probabilistic bias analysis and Bayesian approaches in simulation studies to address mean squared error (MSE) and 95% credible interval width (the difference between the log-OR confidence intervals). We performed two sets of simulations, one in which the distribution for sensitivity among cases was close to the admissibility bounds and the other in which it was further away. For both simulations, we specified true exposure prevalences as r0=0.45 and r1=0.50, implying ψ=1.2. Further, we assumed q0=0.95, q1=0.70, and p0=0.9 in both simulations. Finally, in the first simulations we specified p1=0.60, while in the second set of simulations we specified p1=0.80. Note that θ1=0.45 in the first set of simulations and θ1=0.55 in the second set. For each set of simulations we generated data for 1000 cases and 1000 controls.
We chose highly informative prior distributions for q0, q1, and p0, and explored the results of the analyses under three prior specifications for p1. We used the distributions described in expressions (2) and (3), and we chose parameter values such that the prior on the sensitivity in cases can be thought of as vaguely, weakly or highly informative. We specify a0=logit(0.9), a1=logit(0.6), , and ρp=0.8. We refer to this prior as vaguely informative in the sense that it states we are 95% certain the true sensitivity among cases is between 0.05 and 0.98, with a best guess of 0.6. As in the applied example, our intention is not to suggest such a vague prior is defensible, but to detect points of divergence between probabilistic bias analysis and Bayesian analyses. The weakly informative prior specifies , with other parameters kept at the same values. This implies we are 95% certain the true sensitivity lies between 0.17 and 0.91. Finally, the highly informative prior specifies , implying we are 95% certain the true value lies between 0.45 and 0.74.
In the second set of simulations (where the true value of p1= 0.80), we define similar vaguely ( ), weakly ( ) and highly ( ) informative priors for this set of simulations with a1=logit(0.8). To complete the specification of the models in both simulations, prior distributions were specified for the specificity parameters: b0=logit(0.95), b1=logit(0.7), , and ρq=0.8. These parameters for specificity are “correctly” specified in the sense that they are centered around the truth with modest uncertainty.
In each of the 6 simulations (2 true sensitivity values × 3 prior specifications), we generated 100 datasets. Each dataset was analyzed using probabilistic bias analysis and Bayesian approaches. Each probabilistic bias analysis was run for 10,000 iterations and each Bayesian analysis was run for 10,000 iterations (following a 1000-iteration burn-in phase). Because resulting distributions tended to have a very heavy right skew, we opted to base inferences on the median rather than the mean. Point estimates and highest posterior density intervals were computed for each simulation. We compared the four approaches under the different prior specifications by mean squared error (MSE) as well as the average interval width (upper 95% CI for the OR minus the lower 95% CI for the OR).
Results from the simulation study are given in Table 2. For the set of simulations in which the true p1 is close to the admissibility bounds, the Bayesian approach had reduced MSE when a vaguely or weakly informative prior was used. With a highly informative prior, no difference was seen in MSE. In the set of simulations in which the true p1 is further from the admissibility bounds, the Bayesian approach was observed to have an increased MSE relative to the probabilistic bias analysis approaches when an uninformative prior was used. With weakly or highly informative prior distributions, no difference in MSE was observed.
Table 2.
Mean squared errors of the OR and CI width for the OR from simulation study of the probabilistic bias analysis and Bayesian approaches to correcting for misclassification.
| Truth: p1=.6 | Truth: p1=.8 | |||
|---|---|---|---|---|
| Bayes | Probabilistic bias analysis | Bayes | Probabilistic bias analysis | |
| Vaguely Informative | ||||
| MSE | 0.14 | 0.40 | 0.02 | 0.07 |
| Widtha | 12.3 | 5.1 | 11.4 | 5.9 |
| Weakly Informative | ||||
| MSE | 0.04 | 0.19 | 0.04 | 0.03 |
| Width a | 14.6 | 6.9 | 10.4 | 5.8 |
| Highly Informative | ||||
| MSE | 0.09 | 0.07 | 0.03 | 0.03 |
| Width a | 7.0 | 4.4 | 1.6 | 1.4 |
The difference between the upper and lower bounds of the 95% CI for the OR
The average posterior interval widths are also given in Table 2. Posterior interval widths can be quite large in uncertainty analyses, and this is reflected in the results. Vaguely and weakly informative priors resulted in wide posterior intervals. We note that scenarios in which the probabilistic bias analysis and Bayesian approaches result in substantially different MSE are often (although not always) accompanied by particularly wide posterior intervals.
Discussion
A careful comparison of the probabilistic bias analysis and Bayesian approaches to exposure misclassification in case-control studies indicates that the two methods frequently perform equally well. In many situations, the results of a probabilistic bias analysis can be interpreted explicitly as Bayesian results with a uniform prior on r0 and r1. Situations do exist in which probabilistic bias analysis should not be viewed as an approximately Bayesian approach and can perform poorly. Fortunately, these situations are relatively easy to detect, and may involve unrealistic prior specifications.
Probabilistic bias analysis represents a Bayesian approach in which the prior distribution of the bias parameter is not correctly updated with the observed data. The data contain somewhat limited information to update the prior distributions, so this inaccurate updating may be relatively inconsequential. The admissibility bounds given in the Appendix restrict the conditional posterior bias parameter distributions to certain regions of the sample space, once the data are observed. If the prior distributions for the bias parameters are almost completely contained within the bounds, then the data have nothing to contribute to the estimation of that bias parameter, and the prior and posterior distributions will look nearly identical. In this case, probabilistic bias analysis is a nearly perfect approximation of a Bayesian analysis. However, if a non-negligible amount of the prior distribution falls outside of the admissibility bounds, then the data can inform the bias parameter distribution. In this case, the probabilistic bias analysis results may not be a suitable approximation of the Bayesian approach, and interpretation of the probabilistic bias analysis may be questionable.
Our simulation results indicate that the Bayesian approach can have lower mean squared errors when substantial portions of the bias parameter prior distribution fall outside the admissibility bounds. It is important to note that in these cases, posterior bias parameter distributions in a Bayesian analysis tend to allocate more probability density to values of that bias parameter that result in a higher variance for the adjusted OR. For instance, in our applied example, under a vaguely informative prior, sensitivity values near 0.26 are favored relative to probabilistic bias analysis. These values of sensitivity imply very few unexposed cases, resulting in a markedly increased variance and wide intervals.
By calculating the probability that the prior distribution falls outside the admissibility bounds, an analyst can easily check whether probabilistic bias analysis is likely to be a decent approximate Bayesian analysis. If the total probability is larger than a few percent, the approximation may be poor. However, in the event that probabilistic bias analysis is a poor approximation of the Bayesian result, our simulations indicate that this is exactly the scenario in which no adjustment method is likely to be very useful the posterior interval width for the adjusted OR is so large as to make the results relatively useless. Upon seeing that a non-negligible portion of the prior probability falls in the inadmissible region, it may be tempting to alter one’s prior so that this did not happen. Such tampering with a prior distribution in light of the observed data is to be discouraged; such a procedure would have unknown properties.
We have repeatedly focused on the distribution of sensitivity or specificity conditional on θ0, θ1. This was to aid in our comparison with the probabilistic bias analysis approach that also views these parameters conditionally. Conditionally, there are inadmissible values of sensitivity and specificity; however, because θ0, θ1 are random and vary from iteration to iteration, all values of sensitivity and specificity are admissible marginally (albeit perhaps very unlikely). Further, in our Bayesian approach, we have assumed a uniform prior on r0 and r1 to aid in our comparison with probabilistic bias analysis. In many instances, epidemiologists may wish to incorporate substantive information on the exposure-disease relationship. Greenland’s data augmentation approach may be the most practical in such instances.18,19 However, specifying informative priors on the exposure probabilities may be quite difficult, in that prior knowledge about these probabilities may be based on studies that also experience exposure misclassification.
Misclassification is a common problem in observational research. Probabilistic bias analysis and Bayesian techniques are methods to adjust effect estimates for misclassification errors. Probabilistic bias analysis has an appealing advantage over the Bayesian approach as it is much easier to implement. The extent to which these various techniques mimic one another has remained unknown. Further, without a formal theoretical framework (Bayesian or frequentist, for example) the interpretation of probabilistic bias analysis results has remained problematic. The results of our research indicate that probabilistic bias analysis is a very useful, often nearly exact, approximation of a Bayesian analysis.
Supplementary Material
Acknowledgments
Funding source: Richard MacLehose was supported by a grant from the National Institutes of Health (1U01-HD061940).
We thank Sander Greenland and Jay Kaufman for very helpful reviews of an earlier draft.
Appendix: Bounds for admissible sensitivities and specificities
The bounds for admissible sensitivities and specificities are identical in the probabilistic bias analysis and Bayesian approaches. We develop them by noting that the prevalence of true exposure must fall between zero and 1: 0 ≤ ri ≤ 1. Noting that the relationship between the true and apparent exposure probabilities is: θi = ri pi+ (1−ri)(1− qi). Inverting this equation, we see the following must hold: . Isolating θi we find the following bounds: . Alternatively, we could solve the inequality for pi and qi to obtain the following bounds:
Footnotes
SDC Supplemental digital content is available through direct URL citations in the HTML and PDF versions of this article (www.epidem.com).
References
- 1.Greenland S. Accounting for uncertainty about investigator bias: disclosure is informative: How could disclosure of interests work better in medicine, epidemiology and public health? Journal of Epidemiology and Community Health. 2009;63(8):593–598. doi: 10.1136/jech.2008.084913. [DOI] [PubMed] [Google Scholar]
- 2.Maclure M, Schneeweiss S. Causation of bias: the episcope. Epidemiology. 2001;12(1):114–22. doi: 10.1097/00001648-200101000-00019. [DOI] [PubMed] [Google Scholar]
- 3.Cornfield J, Haenszel W, Hammond EC, Lilienfeld AM, Shimkin MB, Wynder EL. Smoking and lung cancer: recent evidence and a discussion of some questions. J Natl Cancer Inst. 1959;22(1):173–203. [PubMed] [Google Scholar]
- 4.Flanders WD, Khoury MJ. Indirect assessment of confounding: graphic description and limits on effect of adjusting for covariates. Epidemiology. 1990;1(3):239–46. doi: 10.1097/00001648-199005000-00010. [DOI] [PubMed] [Google Scholar]
- 5.MacLehose RF, Kaufman S, Kaufman JS, Poole C. Bounding causal effects under uncontrolled confounding using counterfactuals. Epidemiology. 2005;16(4):548–55. doi: 10.1097/01.ede.0000166500.23446.53. [DOI] [PubMed] [Google Scholar]
- 6.Greenland S, Lash TL. Bias analysis. In: Rothman KJ, Greenland S, Lash TL, editors. Modern Epidemiology. 3. Philadelphia: Lippincott-Williams-Wilkins; 2008. pp. 345–380. [Google Scholar]
- 7.Eddy DM, Hasselblad V, Shachter RD. Statistical modeling and decision science. Boston: Academic Press; 1992. Meta-analysis by the confidence profile method : the statistical synthesis of evidence. [Google Scholar]
- 8.Greenland S. Multiple-bias modelling for analysis of observational data. Journal of the Royal Statistical Society, A. 2005;168(2):1–25. [Google Scholar]
- 9.Lash TL. Applying quantitatvie bias analysis to epidemiologic data. New York: Springer; 2009. [Google Scholar]
- 10.Lash TL, Fink AK. Semi-automated sensitivity analysis to assess systematic errors in observational data. Epidemiology. 2003;14(4):451–8. doi: 10.1097/01.EDE.0000071419.41011.cf. [DOI] [PubMed] [Google Scholar]
- 11.Phillips CV. Quantifying and reporting uncertainty from systematic errors. Epidemiology. 2003;14(4):459–66. doi: 10.1097/01.ede.0000072106.65262.ae. [DOI] [PubMed] [Google Scholar]
- 12.Gustafson P. Measurement error and misclassification in statistics and epidemiology : impacts and Bayesian adjustments. Boca Raton: Chapman & Hall/CRC; 2004. [Google Scholar]
- 13.Gustafson P, Le N, Saskin R. Case-control analysis with partial knowledge of exposure misclassification probabilities. Biometrics. 2001;57:598–609. doi: 10.1111/j.0006-341x.2001.00598.x. [DOI] [PubMed] [Google Scholar]
- 14.Fox MP, Lash TL, Greenland S. A method to automate probabilistic sensitivity analyses of misclassified binary variables. Int J Epidemiol. 2005;34(6):1370–6. doi: 10.1093/ije/dyi184. [DOI] [PubMed] [Google Scholar]
- 15.Chu HT, Wang ZJ, Cole SR, Greenland S. Sensitivity analysis of misclassification: A graphical and a Bayesian approach. Annals of Epidemiology. 2006;16(11):834–841. doi: 10.1016/j.annepidem.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 16.MacLehose RF, Olshan AF, Herring AH, Honein MA, Shaw GM, Romitti PA, Stud NBDP. Bayesian Methods for Correcting Misclassification An Example from Birth Defects Epidemiology. Epidemiology. 2009;20(1):27–35. doi: 10.1097/EDE.0b013e31818ab3b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steenland K, Greenland S. Monte Carlo sensitivity analysis and Bayesian analysis of smoking as an unmeasured confounder in a study of silica and lung cancer. Am J Epidemiol. 2004;160(4):384–92. doi: 10.1093/aje/kwh211. [DOI] [PubMed] [Google Scholar]
- 18.Greenland S. Bayesian perspectives for epidemiologic research III. Bias analysis via missing-data methods. International Journal of Epidemiology. 2009;38:1662–1673. doi: 10.1093/ije/dyp278. [DOI] [PubMed] [Google Scholar]
- 19.Greenland S. Relaxation penalties and priors for plausible modeling of nonidentified bias sources. Statistical Science. 2009;24(2):195–210. [Google Scholar]
- 20.Greenland S. Variance estimation for epidemiologic effect estimates under misclassification. Stat Med. 1988;7(7):745–57. doi: 10.1002/sim.4780070704. [DOI] [PubMed] [Google Scholar]
- 21.Honein MA, Rasmussen SA, Reefhuis J, Romitti PA, Lammer EJ, Sun L, Correa A. Maternal smoking and environmental tobacco smoke exposure and the risk of orofacial clefts. Epidemiology. 2007;18(2):226–33. doi: 10.1097/01.ede.0000254430.61294.c0. [DOI] [PubMed] [Google Scholar]
- 22.Klebanoff MA, Levine RJ, Clemens JD, DerSimonian R, Wilkins DG. Serum cotinine concentration and self-reported smoking during pregnancy. Am J Epidemiol. 1998;148(3):259–62. doi: 10.1093/oxfordjournals.aje.a009633. [DOI] [PubMed] [Google Scholar]
- 23.Klebanoff MA, Levine RJ, Morris CD, Hauth JC, Sibai BM, Ben Curet L, Catalano P, Wilkins DG. Accuracy of self-reported cigarette smoking among pregnant women in the 1990s. Paediatr Perinat Epidemiol. 2001;15(2):140–3. doi: 10.1046/j.1365-3016.2001.00321.x. [DOI] [PubMed] [Google Scholar]
- 24.Pickett KE, Rathouz PJ, Kasza K, Wakschlag LS, Wright R. Self-reported smoking, cotinine levels, and patterns of smoking in pregnancy. Paediatr Perinat Epidemiol. 2005;19(5):368–76. doi: 10.1111/j.1365-3016.2005.00660.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


