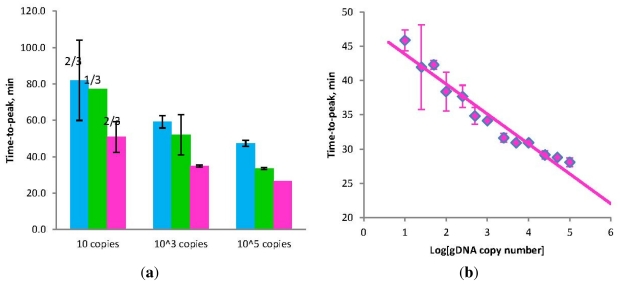
Figure 2.
(a) Amplification times for LAMP without Loop primers (blue) and STEM-LAMP with Stem primers instead of Loop primers (pink) for 10, 1000 and 100,000 copies of genomic Clostridium difficile DNA target; STEM-LAMP without displacement primers for the same number of copies is shown in green; (b) a six log dilution series of Clostridium difficile toxin B gene using STEM-LAMP (all dilutions were measured and detected in triplicate unless indicated otherwise). Final primers concentrations were: Cd-BIP, Cd-FIP, Cd-StemB and Cd-StemF—1.6 μM each, Cd-DisplB and Cd-DisplF—0.4 μM each. Each set of data included three no template controls (NTCs), that remained clean throughout the duration of the assay and were not associated with any time-to-peak value (hence not shown graphically). Error bars represent standard deviations of triplicate measurements.