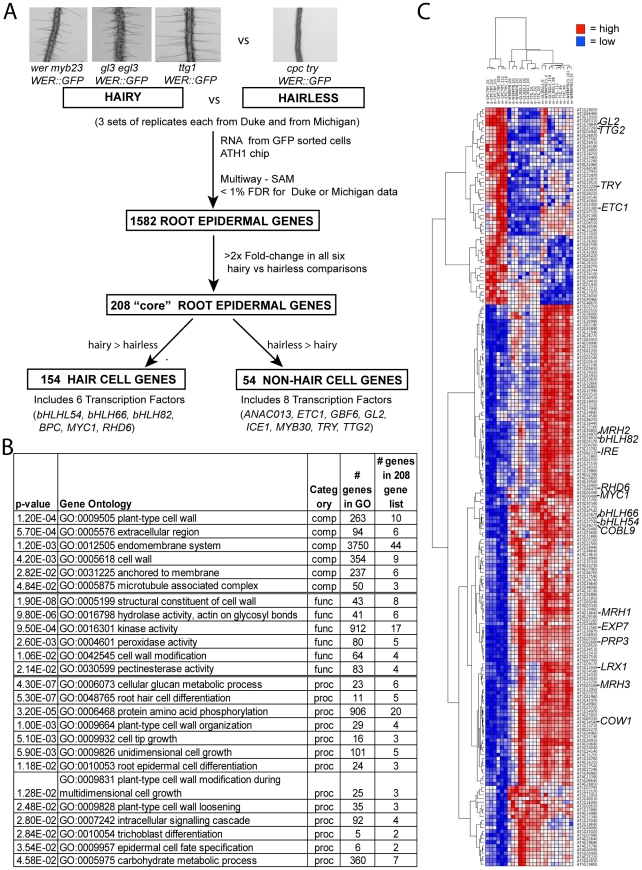
Figure 2. Identification of genes in the root epidermis differentiation pathway.
(A) Flow chart of the steps used to define 154 root-hair cell genes and 54 non-hair cell genes used to build the root epidermal gene network. (B) List of gene ontology (GO) categories that are significantly (p<0.05) overrepresented among the core 208 root epidermal genes. (C) Hierarchical clustering of the 208 core root epidermal genes, based on their relative transcript accumulation in Affymetrix ATH1 microarrays using WER::GFP-expressing cells from three replicates of the wer myb23, gl3 egl3, ttg, and cpc try mutants. Red = high transcript level; Blue = low transcript level. The order of microarray samples along the x-axis is as follows: Columns 1–3, cpc try (Duke); Columns 4–6, cpc try (Mich); Columns 7–9, wer myb23 (Duke); Columns 10–12, gl3 egl3 (Duke); Columns 13–15, ttg (Duke), Column 16, gl3 egl3 (Mich); Column 17, wer myb23 (Mich); Columns 18–19, gl3 egl3 (Mich); Columns 20–22, ttg (Mich); Columns 23–24, wer myb23 (Mich). On the right side, specific gene names represent the genes analyzed in the mutant microarrays or genes previously known to be regulated by the WER/MYB23-GL3/EGL3-TTG pathway.