Figure 5. Structural model refinement of the transmembrane domain for the membrane-bound fd-coat protein.
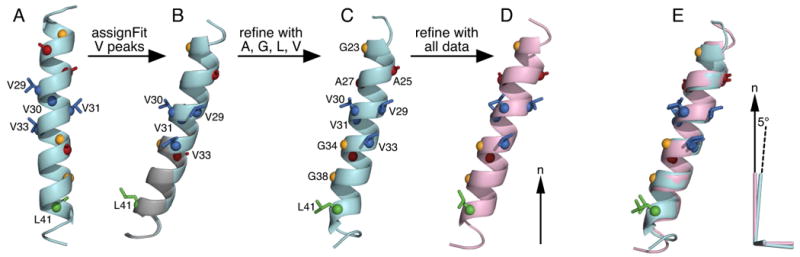
Coordinates are oriented in the frame of the lipid bilayer, relative to the lipid bilayer normal (n; arrow). (A) Ideal helix structural model (arbitrary orientation) used to initiate the SASR cycle. (B) Rigid body orientations of the starting structural model obtained for the lowest (cyan) and second lowest (gray) RMSDtot AssignFit assignment of the Val peaks. Swapping the assignments of V29 and V33 has little effect on model orientation. (C) Structure obtained after refinement with the DC and CSA restraints from Leu, Val, Ala and Gly. (D) Structure obtained after refinement with the DC and CSA restraints from all of the previously measured DC and CSA restraints [18]. (E) The AGLV-refined structure (cyan) is aligned to the all-data-refined structure (pink). The cyan and pink axes represent the order tensors for the respective structures. The pink principal axis coincides with the lipid bilayer normal, while the cyan principal axis is off by 5°. Alignment was obtained for the backbone Cα atoms.
