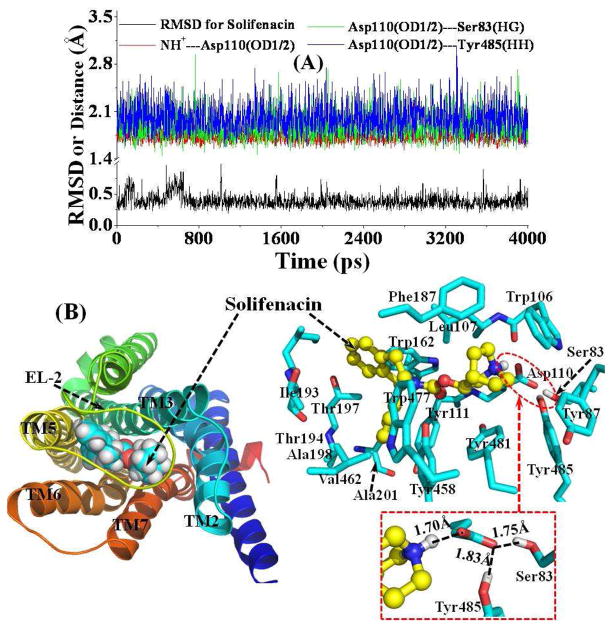
Figure 4.
(A) Plots of the tracked positional RMSD for Solifenacin from its original coordinates and tracked distances. NH+---Asp110(OD1/2) represents the shortest distance between the proton at the cationic head of Solifenacin and the negatively charged atoms (OD1 or OD2) on the side chain of residue Asp110; Asp110(OD1/2)---Ser83(HG) represents the distance for the hydrogen bond formed by the Asp110 side chain and the Ser83 side chain; Asp110(OD1/2)---Tyr485(HH) is the distance for the hydrogen bond formed by the Asp110 side chain and the Tyr485 side chain. (B) Top view of representative structure for M5 mAChR-Solifenacin complex, taken from the last snapshot of the MD simulations. The coloring scheme for the complex structure is the same as that used in Figure 2B, and the Solifenacin is presented as spheres (left panel) or ball-and-stick (right panel). Residues within 5 Å of Solifenacin are labeled and shown in stick. The hydrogen bonding interaction between the Asp110 side chain and the cationic head of Solifenacin, and the hydrogen bonding interactions among side chains of Asp110, Ser83, and Tyr485 are shown in dashed lines along with the labeled distances.