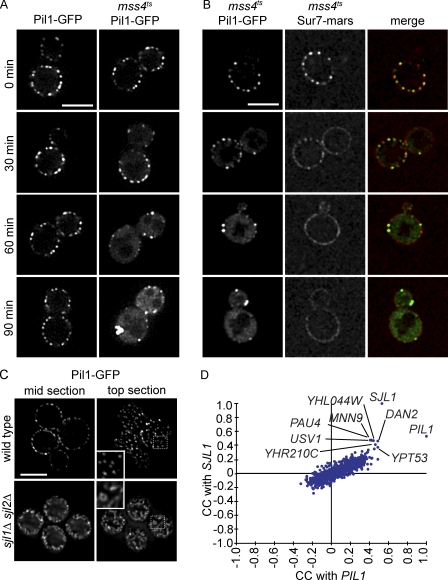
Figure 3.
PI(4,5)P2 is necessary for normal eisosomes in vivo, and PIL1 has a highly similar genetic profile to SJL1. (A) Fluorescence microscopy of Pil1-GFP in a yeast mutant strain containing a temperature-sensitive allele of MSS4 (mss4ts). Pil1-GFP loses its normal eisosome pattern but instead clusters to enlarged structures at the membrane after a 90-min (right column) temperature shift from 24 to 37°C. The control strain does not show this phenotype (left column). (B) Fluorescence microscopy of Sur7-mars and Pil1-GFP in mss4ts cells. After 30 min of temperature shift, Sur7-mars loses its localization to the MCC. After 60 min, it is evenly distributed in the plasma membrane. (C) Deletion of SJL1 and SJL2 results in increased Pil1-GFP assembly at the plasma membrane. Insets show magnified regions of cells in the boxed areas. Bars, 5 µm. (D) Comparison of correlation scores from an E-MAP focusing on lipid metabolism. SJL1, encoding the PI(4,5)P2 phosphatase, has the most similar genetic signature to PIL1, indicating similar gene function. CC, correlation of correlations.