Abstract
Research in cell biology and the development of translational technologies are driven by competition, public expectations, and regulatory oversight, putting these fields at a critical juncture. Success in these fields is quickly becoming dependent on the ability of researchers to identify and isolate specific cell populations from heterogeneous mixtures accurately and efficiently. Many methods for cell purification have been developed, and each has advantages and disadvantages that must be considered in light of the intended application. Current cell separation strategies make use of surface proteins, genetic expression, and physics to isolate specific cells by phenotypic traits. Cell purification is also dependent on the cellular reagents available for use and the intended application, as these factors may preclude certain mechanisms used in the processes of labeling and sorting cells.
Key Words: Cell purification, separation, sorting; Virus; Recombineering
The Unresolved Problem of Cell Purification
Advances in cell biology and tissue engineering have expanded the horizons toward which researchers and clinicians can look; however, in many cases, the implementation of cell-based technologies faces the same logistical and regulatory hurdles that it has always faced. As is the nature of science, when the bar is raised for what is possible it is quickly followed by the bar being raised for what is expected, both by study sections and the general populace. If a paper reveals an interesting phenotype in an animal model today, tomorrow's grants will need to propose isolation of the responsible mechanism. In many ways, today's science is a race to make current technologies and techniques obsolete. It is interesting, therefore, that the problem of cell purification has persisted for so long. The relevance of conclusions made in basic science research and the utility of cell-based technologies hinge upon our ability to reliably and consistently identify and subselect individual cell populations [Szaniszlo et al., 2004], yet investigators still struggle to accomplish this fundamental step.
One of the most essential goals of research conducted within the basic sciences is to maximize the confidence that the cell models employed are representative of physiological realities. Mixed and misidentified cell populations introduce uncertainty with respect to the mechanistic causes of phenomena observed in experimental models. Cardiovascular research is plagued by these uncertainties as pure populations of individual cell types found in the heart are difficult to obtain. Even in the lucky case where the cell type under study has been researched well enough that a unique, validated marker profile has been identified, contaminating cells, such as fibroblasts, will continue to hinder mechanistic experiments unless purification techniques are applied. By developing and employing reliable methods of cell purification, it is possible to exert much more control over these aspects of biological research. This, in turn, improves both the confidence and the reproducibility of published results while minimizing the introduction of experimental discrepancies between laboratories.
Accurate cellular identification and purification is even more crucial in translational applications. Devices and technologies designed for use in a clinical setting are subjected to strict review by governmental oversight agencies [Simek, 2005; Arcidiacono et al., 2010; Nelson et al., 2010] before they can be brought to market. During this process, the causative cellular agent must be identified to ensure that the clinical outcome can be specifically attributed to the efficacy of the cell-based device [Simek, 2005; Nelson et al., 2010]. Furthermore, patent law in the USA is at a critical juncture that will have serious ramifications for the use of engineered tissues and organs [Harris, 2008]. While patents have been issued for stem and progenitor cell populations based on surface marker expression [Huang, 2010], it remains to be seen how rulings will be made regarding cells that have been introduced into the body. An example in which cell identification and purification become central issues in bioengineering is that of a cell population expressing one patented set of markers that is found to express another patented set of markers after injection or implantation. It is not clear how the legal specifics of issues like this one will be resolved, but it is apparent that unerring control over cell population purity will be essential for the development of cell-based therapeutics.
Purification Using Cell Surface Proteins
In general, it is desirable to employ the least invasive method available to isolate and sort cells for use in experimental biology or bioengineering. In many cases, the easiest way to accomplish these goals is to utilize cells’ surface proteins (fig. 1a), based on either expression or functionality, to purify a subpopulation from heterogeneous cell mixtures using fluorescence-activated cell sorting (FACS), magnetic sorting, or adhesion-based cell separation. Some commonly used surface markers for cardiac progenitor cell purification are cellular prion protein, SSEA1, and KDR [Yang et al., 2008; Blin et al., 2010; Hidaka et al., 2010]. Hyperpolarization-activated cyclic nucleotide-gated potassium channel 4 (HCN4) and elastin microfibril interface 2 (EMILIN2) are surface proteins that can be used to specifically label differentiated cardiac nodal cells and cardiomyocytes, respectively [Dobrzynski et al., 2003; Van Hoof et al., 2010]. Conversely, surface protein-based cell purification can also be used in negative selection strategies like complement depletion to deplete a population of undesired cell types. Surface protein-based sorting is a powerful tool in many research projects as it is considered minimally disruptive to cell phenotype and viability, but it is not without its limitations and caveats (table 1).
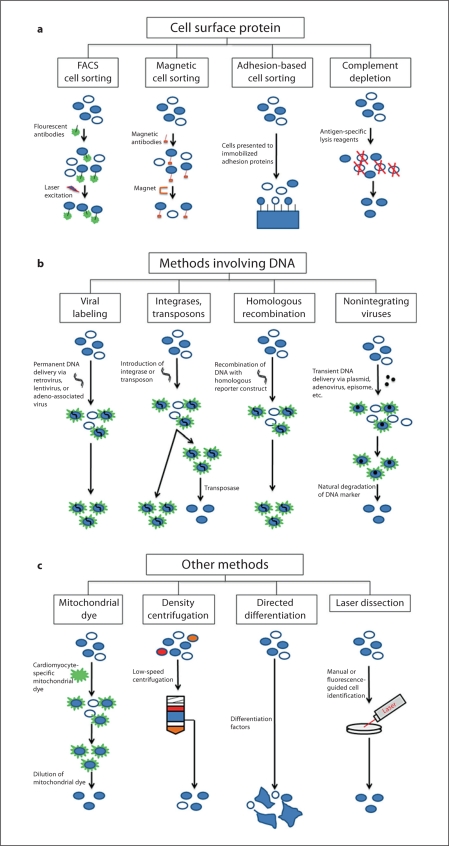
Fig. 1.
Methods of cell purification. a Purification methods contingent on the use of surface markers. b Purification methods contingent on the use of DNA. c Other methods used to purify cells.
Table 1.
Summary of advantages and disadvantages of current cell purification techniques
| Sorting method | Advantages | Disadvantages | First author, year |
|---|---|---|---|
|
|
|
|
| Magnetic cell sorting |
|
|
Nohe, 2002; Zborowski, 1996 |
| Adhesion-based cell sorting |
|
|
Lu, 2010; Cavallo, 2010; Diaz-Prado, 2010; Freund, 2010; Yona, 2010; Adams, 2010 |
| Complement depletion | Accurate | Cell lysis may adversely affect other cells | Dittel, 2010 |
| Viral vectors |
|
|
Lu, 2004; Kimatcheva, 1999; Fu, 2010; Philpott, 2007 |
| Integrases and transposons |
|
|
Shibano, 2007; Mejia-Pous, 2009; Tenzen, 2010 |
| Homologous recombination |
|
|
Jonkers, 2002; Tian, 2006; Davison, 2008; Yang, 2008 |
| BAC recombineering |
|
Sarov, 2006; Song, 2010 | |
| Zinc finger recombineering | Improved efficiency over BAC | Difficult to design endonucleases | Bobis-Wozowicz, 2010; Cui, 2010 |
| Nonintegrating vectors | |||
| Plasmids | Transient expression of reporter | Very low efficiency | Okita, 2008 |
| Episomes | Transient expression of reporter |
|
Yu, 2009 |
| Adenovirus |
|
Viral | Hermening, 2006 |
| Mitochondrial dye |
|
|
Hattori, 2010 |
| Density purification | Fast and inexpensive |
|
Xu, 2006; Horn, 2007; Zhang, 2010 |
| Purification through differentiation of progenitor cells |
|
|
Mummery, 2003; Nemir, 2006; Yuasa, 2005; Laflamme, 2007; Ieda, 2010 |
| Laser dissection |
|
|
Koller, 2004; Szaniszlo, 2006; Hempel, 2007 |
The strengths and weaknesses of each purification method must be considered when designing experiments for cell biology and cell-based therapeutics. For example, cell sorting based on surface protein expression is more amenable to translational projects since it does not require permanent manipulation of the cell, but expression of these proteins may not be as precise as the expression of genes in the nucleus.
Surface Protein-Based FACS
The most prevalent method of cell purification is FACS [Ebbe, 2000]. The popularity of FACS is largely due to its significant efficiency and accuracy. FACS is widely used to separate particular cell populations for use as reagents in both basic science and translational experiments. The method uses phenotype-specific surface markers in conjunction with fluorescent antibodies to those markers. The cells to be purified are then presented to a fluorescent excitation laser and separated based on the presence or absence of one or more fluorescently labeled cell surface proteins. FACS plays a critical role in several other purification strategies entailing induction of intracellular fluorescent markers, such as green fluorescent protein (GFP), in which the need for protein-specific antibodies is obviated. A complementary functionality of most FACS equipment is the use of forward- and side-scatter optics to differentiate cells based on size and internal complexity. While these functions are far less specific than the antibody-based FACS functions, they can be useful as an additional means of discerning cell types and assessing viability [Ohnuma et al., 2006].
The costs associated with FACS equipment often prohibit individual labs from owning their own FACS equipment; however, the general utility of the technique usually justifies interdepartmental FACS core facilities at most institutions. This process is relatively high throughput, but the sorting functionality becomes impractical with small sample sizes [Metezeau, 1993]. The accuracy of FACS is dependent on the ratio of sample pressure to sheath pressure and the number of markers being used to differentiate between populations. A higher sample-to-sheath pressure ratio can be used to sort large numbers of cells quickly, but collected cell viability and the accuracy of cell sorting are decreased slightly. This is due to an infrequent occurrence in which two cells pass in front of the detectors at the same time. Similarly, overall sorting accuracy decreases with each additional marker used to define a cell population.
One of the major drawbacks of FACS is that it is limited to surface marker protein detection. In some applications, especially those related to developmental biology and tissue engineering, many of the proteins considered critical to discriminating one cell population from another are intracellular and cannot easily be labeled with antibodies in live cells. Also, certain cell types may be too sensitive to mechanical stimulation to survive the FACS sorting process [Schindler et al., 1985; Metezeau, 1993]. Furthermore, it is often difficult to identify surface proteins unique to cells of a certain phenotype or developmental stage. Thus, surface protein-based FACS is only useful if one can identify differential expressions of cell surface markers. Consequently, the process of identifying characteristic surface protein expression profiles is a labor-intensive endeavor that can result in disagreement [Martin-Puig et al., 2008; Schauwer et al., 2011].
Magnetic Cell Sorting
Magnetic cell sorting also utilizes antibody specificity to separate defined cellular subpopulations from larger mixtures, but the antibodies to surface proteins are associated with magnetic conjugates instead of fluorophores. Thus, inexpensive and easy-to-use equipment employing an electromagnet or permanent magnet can be used to retain cells that express the surface marker of interest at a fraction of the cost of the equipment required for FACS. Even though regular magnetic sorting does not cause phenotypic changes by itself [Nohe et al., 2002], it is possible to activate surface protein-mediated phenotypic changes intentionally by applying mechanical force to surface proteins via bound magnetic beads [Lew et al., 1999]. The drawbacks of surface protein-based purification technology are felt more acutely when using magnetic sorting techniques since there are fewer antibodies available for magnetic cell sorting than FACS and only one marker can be sorted at a time in magnetic cell sorting. Also, it has been reported that the accuracy of magnetic cell sorting is not as high as that of FACS with respect to false positives (largely due to cell-cell adhesion) and false negatives [Zborowski et al., 1996]. As magnetic sorting is useful for bulk enrichment but is not as precise as FACS, it is unlikely that magnetic sorting, on its own, can be improved to produce sufficiently pure populations for direct use in clinical science.
Adhesion-Based Cell Sorting
FACS and magnetic cell sorting are both very useful technologies, but they are, in essence, predicated on protein expression instead of a functional readout. In certain applications, such as studies involving circulating cells [Lu et al., 2010] or unpurified mesenchymal stem cell samples [Diaz-Prado et al., 2010; Freund et al., 2010; Cavallo et al., 2011], it can be useful to employ adhesion as a selection criterion for subpopulation sorting. Monocytes have been shown to play a role in the regeneration of postinfarction myocardium [Zuidema and Zhang, 2010], so the use of microprinted or adsorbed adhesion or matrix proteins can be useful in subselecting the populations of most interest [Adams et al., 2008; Yona et al., 2010]. Similarly, much of today's adult progenitor research involving mesenchymal stem cells incorporates a distinction between adherent and nonadherent cell populations [Nomura et al., 2007; Diaz-Prado et al., 2010; Freund et al., 2010; Jing et al., 2010]. The techniques used for adhesion-based cell sorting can be fast and inexpensive, but they can also be confounded by the fact that many adhesion processes simultaneously involve multiple adhesion proteins and mechanisms [Freund et al., 2010; Pamukcu et al., 2010]. Therefore, some disadvantages of adhesion-based cell sorting are sorting efficiency that depends on the methodology and false positives as a result of cell-cell interactions between cells of interest and contaminating cells in the mixture.
Complement Depletion
An alternative approach to population purification called complement depletion uses monoclonal antibodies to destroy specific cells from a mixed population. Briefly, this technique uses activated plasma proteins to create pores in targeted cells’ surfaces through a proteolytic cascade [Dittel, 2010]. These cells subsequently lyse and are washed from the sample. The process is highly efficient (95–100%) at destroying particular cell populations. Most researchers with access to FACS equipment would likely forgo complement depletion as FACS would offer a more reliable system for selective depletion; nevertheless, complement depletion offers an interesting and potentially useful approach to cell purification.
Purification Methods Involving DNA Sequences
While surface proteins are the most accessible means of discerning one cell type from another, issues with antibody technology, including specificity, availability, and cross-reactivity, leave room for improvement. A number of alternative strategies have been developed to circumvent the limitations of antibodies. The two most widely used gene modulation techniques for cell purification are to induce expression of sortable markers or to confer resistance to selection agents [Muller et al., 2000; Tian et al., 2006; Davison et al., 2008]. These techniques can be combined to give both positive and negative selection [Jonkers and Berns, 2002; Tian et al., 2006; Huber et al., 2007; Davis et al., 2008], further improving the efficiency of cell type isolation. The transgenic expression can also be made by use of a minimal promoter and a condition-specific enhancer sequence [Muller et al., 2000; Jonkers and Berns, 2002; Tian et al., 2006; Huber et al., 2007; Liu et al., 2010]. Some common markers of cardiac phenotype that are difficult to label in living cells with antibodies are myosin light chain 2a (mlc2a), myosin light chain 2v (mlc2v), NK2 transcription related factor locus 5 (Nkx2.5), atrial natriuretic peptide (ANP), connexin 43, α-actinin, cardiac troponin I (cTnI), and cardiac troponin T (cTnT) [Kubalak et al., 1994; Chen et al., 1998; Muller et al., 2000; Huber et al., 2007; Bu et al., 2009; Fu et al., 2010]. The question then becomes the best means of introducing the desired selection sequence(s). Four main strategies have been developed to address this question: viral infection, transposase recombination, homologous recombination, and zinc finger recombination (fig. 1b).
Viral Labeling
The most straightforward means of inserting a genetic sequence into a cell's genome is to make use of the mechanisms that viruses have optimized over billions of years. By introducing the desired genetic reporter sequence along with the necessary viral sequences into a host cell type (such as HEK 293T cells), one can produce a large number of viruses capable of delivering the reporter sequence to a cell population. Cells expressing the reporter can then be isolated from the larger heterogeneous mixture using selection techniques like FACS or enrichment through drug resistance.
Retroviral, lentiviral, and adeno-associated viruses can be used to permanently incorporate the reporter sequence into the genome [Klimatcheva et al., 1999; Fu et al., 2010]. However, retroviruses can only infect dividing cells, making infection of slowly dividing cells like neurons potentially problematic [Kalpana, 1999]. Adeno-associated viruses are much smaller than other virus types and can only carry sequences up to 4.8 kb in length, meaning that many reporter sequences are too large to be delivered by adeno-associated viruses [Lu, 2004]. Lentiviruses offer high-efficiency infection of dividing and nondividing cells and a relatively large payload; however, they are similar to retroviruses in that they cause random integration into the genome [Philpott and Thrasher, 2007; Kumar and Woon-Khiong, 2011]. Permanent integration of genetic markers can be very useful in experimental biology, but it represents a roadblock for therapeutic applications, thereby limiting its current utility in the generation of purified cell populations. Recently, systems have been developed which may allow researchers to selectively remove permanently integrated sequences using Cre-loxP following phenotypic sorting [Davis et al., 2008; Moravcikova et al., 2008]. However, even if reporter sequences can be excised, small transgenic sequences remain in the target genome using these systems, presenting the risk of tumor formation.
phiC31 Integrase and Transposons
Two inventive ways of avoiding the size limitations of some viral vectors are to make use of plasmids encoding either integrases or transposases [Tenzen et al., 2010]. The underlying principle of these two technologies is to transiently express an enzyme capable of inserting a synthesized reporter sequence into the genome at specific insertion sites.
Using the phiC31 integrase mechanism observed in bacteria, it is possible to exercise a certain degree of control over the insertion site of reporter sequences. There exist sequences in the mammalian genome that are similar to bacterial attP integration sites [Chalberg et al., 2006]. At these mammalian sites, the same phiC31 integrase responsible for integrating sequences containing an attB site at attP loci in bacteria is able to insert attB-containing reporter sequences [Thyagarajan et al., 2001, 2008]. The number of these insertion sites in the mammalian genome is low, and they are located primarily in introns not known to have transcriptional activity [Tenzen et al., 2010]. Therefore, insertion of reporter sequences using this methodology is unlikely to disrupt endogenous gene transcription. Unfortunately, phiC31 integrase activity is irreversible, so incorporation of reporter sequences with this method is permanent and problematic for projects intended for the clinic.
The transposon system makes use of a large synthesized reporter sequence flanked by inverted transposon recognition sites in combination with a nonintegrating plasmid encoding transposase [Batista Lobo et al., 2007]. The transposase inserts the reporter sequence randomly into the genome, allowing cell sorting based on reporter expression [Shibano et al., 2007; Mejia-Pous et al., 2009]. After cell sorting, another transient transfection with a transposase-encoding vector is used to remove the inserted reporter sequence. Several transposon systems have been used in mammalian models, including Tol2, Sleeping Beauty, Frog Prince, and piggyBAC [Wu et al., 2006]. While the transposon systems like piggyBAC can be removed without any vestiges of transgene in the target cell [Tenzen et al., 2010], the reversible nature of the transposase reaction necessitates an additional sorting step in which cells intended for use in human subjects must be separated based on successful removal of the reporter construct.
Homologous Recombination
Homologous recombination is commonly known in the context of transgenic animal model creation. In the process, a segment of DNA is created containing a reporter and DNA sequences identical or homologous to the gene which is to be targeted. The segment is electroporated into embryonic stem (ES) cells, and, as the cells divide, the homologous regions break, leading to translocation of transgenes and integration into the genome [Jonkers and Berns, 2002; Tian et al., 2006; Davison et al., 2008; Yang et al., 2008; Song et al., 2010]. Successfully labeled cells are selected by drug resistance conferred by the inserted DNA sequence. In the process of creating transgenic animals, ES cells are then injected into allogeneic blastocysts, and the resulting chimeric animals are backcrossed to produce animals expressing the reporter sequence stably. As recombination performed in this manner often utilizes reporter plasmids with regions homologous to a specific gene, expression of particular genes can be quantified throughout an animal by analyzing the reporter signal expressed in place of the gene that was targeted. Compared to viral techniques, homologous recombination provides site-specific integration of the reporter sequence rather than incorporation into random gene sequences. Reporters introduced through homologous recombination are expressed faithfully with respect to the targeted gene, and no transgenic residue is left aside from the inserted reporter cassette. The size of sequences to be inserted using simple homologous recombination is limited, as larger plasmids tend to break and degrade before they can be stably incorporated into the genome. Also, the efficiency of integration during basic homologous recombination in human cells is low. Animals models created using these techniques to express reporter proteins can be used to isolate specific populations of cells.
Another approach to homologous recombination that allows the use of larger sequences involves the use of bacterial artificial chromosomes (BACs). Using BACs, it is possible to insert large reporter sequences and all of the relevant upstream and downstream promoter and enhancer sequences simultaneously. Therefore, BAC technology allows the substitution of a reporter gene into the exact locus of the gene of interest, preserving contextual expression and offering more faithful reporter expression, while increasing the amount of genetic material that can be inserted and increasing the efficiency of integration [Song et al., 2010]. To make the use of homologous recombination and BAC recombination less labor intensive, protocols have been developed to streamline the processes [Sarov et al., 2006]. This allows for commercialization of these technologies and greatly increases their utility in both experimental biology and translational science.
Zinc finger nucleases can be used to improve the efficiency of homologous recombination by significantly increasing the probability of successful double strand breaks in chromosomes, leading to dramatic increases in the likelihood that homologous recombination events will occur [Tenzen et al., 2010; Bobis-Wozowicz et al., 2011; Cui et al., 2011]. Plasmids encoding zinc finger nucleases can be designed to cause breaks in double-stranded DNA at specific loci. Thus, double strand breaks can be introduced into a target cell around a particular gene, and large reporter constructs can be inserted through homologous recombination at an extremely high efficiency without any of the concerns posed by viral approaches. A current drawback of this system is that it is difficult to design custom site-specific zinc finger endonucleases. Without well-designed endonucleases, double strand breaks can be introduced nonspecifically, leading to apoptosis.
Nonintegrating Vectors
Transient transfection with constructs encoding pluripotency genes have been used to induce pluripotency in target cells, but the technology may be amenable to reporter constructs for use in cell purification. In one approach, cells are electroporated and exposed to plasmid vectors which then enter the cells and express the gene(s) that they encode before being degraded by the cell [Okita et al., 2008]. Similarly, episomes have been constructed to encapsulate fabricated plasmid vectors, enhancing vector stability [Yu et al., 2009]. This, in turn, improves the opportunity for the vector to enter the cell and successfully deliver its payload. The efficiency of each method is low, and it may be difficult to incorporate large reporter sequences under the control of cell type-specific promoters.
The capacity of adenoviral vectors normally precludes their ability to deliver large sequences like those needed for use as reporter constructs; however, the advent of high-capacity adenoviruses makes adenoviral technology an interesting option for cell purification [Volpers and Kochanek, 2004]. Adenoviruses do not cause permanent integration into the target cell's genome, and they have been used to transiently deliver tissue-specific gene expression constructs in neurons [Hermening et al., 2006].
Other Purification Methods
Surface proteins and genetic modification are, by far, the most common approaches to sorting one cell type from larger cell mixtures, but other approaches offer options for specialized applications. Many of these alternative techniques can be used in conjunction with surface protein-based sorting or methods involving the introduction of genetic material to increase the yield of highly purified cells (fig. 1c). Thus, these techniques are particularly useful when large quantities of cellular reagents are needed, as is the case for tissue engineering and cell therapy.
Mitochondrial Dye
Recently, Hattori et al. [2010] described a means of specifically labeling cardiomyocytes derived from pluripotent stem cells using a mitochondrial labeling method. In the study, a Nernstian dye, tetramethylrhodamine methyl (TMRM) ester, capable of entering and exiting the mitochondria in accordance with mitochondrial membrane potential was used as a positive selection marker for cardiomyocytes derived from rat, mouse, and human pluripotent stem cells. As cardiomyocytes are the cell type accounting for the largest volume of the heart [Baudino et al., 2006] and the supply of primary cardiomyocytes available for research is extremely limited, this method promises a quick, nongenetic approach to producing highly enriched (more than 99% pure) populations of cardiomyocytes. It also circumvents the need for genetic manipulation but includes a mixture of both atrial and ventricular cardiomyocytes. Also, special attention must be paid to gating of other fluorescent channels during sorting since it was observed that autofluorescence is common in unstained noncardiomyocyte populations [Hattori et al., 2010]. If one's goal is to isolate cardiomyocytes, this method of mitochondrial labeling is highly effective. However, if the goal is to isolate other cardiac cell types such as pacemaker or endocardial cells, new dyes are needed.
Density Purification
Basic cell sorting can be performed based on cell size and density using defined centrifugation media. By creating layers of media with successively lower densities inside of a cell sample and centrifuging at low speeds, it is possible to separate a mixed cell population based on the sizes of its component cell types. Adult progenitor cells from bone marrow are routinely enriched in this manner [Horn et al., 2008; Zhang et al., 2011]. It is also possible to enrich cardiomyocytes from ES-derived populations using density centrifugation, but the resulting population purity is low [Xu et al., 2006; Laflamme et al., 2007]. The method is quick, uncomplicated, and inexpensive, but, used independently, it is relatively inefficient at cell purification and cannot be used to separate individual cell populations of similar size.
Purification through Differentiation of Progenitor Cells
When using progenitor cells as a starting material, one technique for obtaining a pure population of differentiated cells is to guide the cells toward the desired phenotype through manipulation of culture conditions and induction of differentiation. Indeed, there exists a variety of protocols designed to direct progenitor cells toward specified lineages, often using the findings and principals of developmental biology to guide protocol development [Laflamme et al., 2007]. For example, it has been shown that differentiation of human ES cells toward the cardiomyocyte phenotype can be improved through modulation of bone morphogenic protein and Notch signaling or coculture with visceral endoderm [Mummery et al., 2003; Yuasa et al., 2005; Nemir et al., 2006]. These techniques are often relatively inefficient at producing purified populations of the desired cell type on their own, but much of their utility lies in producing large quantities of differentiated cells that can be subsequently purified using methods like FACS or density purification.
More recent advancements in this area have led to the use of direct reprogramming of primary mouse fibroblasts to cardiomyocytes, eliminating the need to use ES cells or create induced pluripotent stem (iPS) cells [Ieda et al., 2010]. This method uses the overexpression of GATA transcription factor 4 (GATA4), myocyte-specific enhancer 2c, and T-box transcription factor 5 to induce a cardiomyocyte phenotype in over 50% of primary fibroblasts. If the method can be improved to incorporate transient, instead of permanent, overexpression of the identified differentiation factors, it provides a valuable tool for incorporation into cardiovascular purification strategies.
Laser Dissection
For applications requiring isolation of primary cells, fluorescence-assisted laser microdissection can be a useful negative selection technique. In laser dissection, computer software is used to guide a medical laser as it ablates nonfluorescent cells in a heterogeneous mixture [Koller et al., 2004; Szaniszlo et al., 2006; Hempel et al., 2007]. When tissue is explanted from transgenic animals harboring fluorescent reporter gene(s), cells expressing the fluorescent reporter gene can be isolated efficiently using laser dissection techniques. Alternately, nonfluorescent cells of interest can be labeled with fluorescent antibodies or microinjected with fluorescent dyes or reporter plasmids and selectively isolated from contaminating cell types. Microinjection can be accomplished using the same laser that will subsequently be used to perform ablation. It has been reported that cell purities in excess of 99% and cell yields of 90% can be obtained from a wide range of sample sizes (from 101 to 108 cells) using in situ laser purification [Koller et al., 2004; Szaniszlo et al., 2006]. It has been shown that laser microdissection can be used to preserve viability in neurons during purification, whereas more widely used techniques like FACS might damage such a fragile cell type [Hempel et al., 2007]. Insofar as it can create highly purified cell populations, laser microdissection has definite utility for isolation of primary cells for use as reagents. At the same time, the necessary laser equipment can be expensive, and the applications for which the technology can be used are limited to those in which cells can be exposed and labeled with fluorescent markers.
Choosing a Cell Source
There are a number of cellular starting materials available for use in the pursuit of biological research. These include: immortal cell lines such as HEK 293, Jurkat, and HeLa cells; pluripotent stem cells such as ES and iPS cells; multipotent adult progenitor cells such as those derived from bone marrow, muscle, skin, fat, and other tissues, and differentiated primary cells, such as keratinocytes, endothelial cells, adipocytes, hepatocytes, and cardiomyocytes. Finding the most effective purification strategy often depends on which and how many cell types are to be incorporated into the experimental system. Similarly, finding the appropriate cell source is dependent on the application for which the cells are to be used.
The ideal situation for studying human biology or engineering therapeutic biological tissues would be to use primary cells in the form of transplanted or explanted human organs and tissues that function in the experimental system similarly to how they do in the host's body. This is frequently infeasible as the quantities of such cells, tissues, and organs available for scientific use are severely limited. Moreover, graft-versus-host interactions can introduce complications in a therapeutic setting, particularly if the tissue is not used autologously. Therefore, researchers must identify the critical parameters of the application in which the cells will be used as each cell source option (differentiated primary cells, multipotent progenitor cells, pluripotent stem cells, and immortalized cell lines) offers a unique set of advantages and disadvantages (table 2).
Table 2.
Comparison of cellular reagents for use in human experimental biology and clinical medicine
| Cell source | Advantages | Disadvantages | Application |
|---|---|---|---|
| Differentiated primary cells |
|
|
|
| Adult multipotent progenitors |
|
|
|
| Pluripotent stem cells |
|
|
|
| Immortal cells |
|
|
|
The selection of cells for use in basic science or translational technologies is often an important decision. Considerations to be contemplated are whether the cells to be used in experimentation are relevant to the physiological equivalent, whether they are plentiful enough for the intended application, and whether they present special concerns like ethical issues.
Differentiated Primary Cells
Differentiated primary cells may be the most obvious choice for use in basic and translational science, but they can also be difficult to use. Some differentiated primary cells such as human umbilical vein endothelial cells (HUVECs) are quite well characterized and can be reliably isolated using well-established protocols [Blackman et al., 2002; O'Connell et al., 2007]. However, HUVECs in an endocardial or blood-brain barrier model may not behave in the same manner as anatomically correct endothelial cells would behave. The derivation of other primary cells can be challenging as isolation protocol development can be arduous. A further limitation of differentiated primary cells is that extended culture leads to a loss of phenotype or cell death [Wolffe and Tata, 1984; Gudjonsson et al., 2004]. This temporal constraint can be devastating to tissue engineering applications, especially for autologous strategies in humans, as it is often infeasible to obtain and expand sufficient quantities of cells for construction implantable products within a small number of population doublings [Vondran et al., 2008; Adam Young et al., 2010].
Multipotent Progenitor Cells
Multipotent progenitor cells have been isolated from a vast number of adult tissues and have been shown to have therapeutic effects in a wide range of applications from bone marrow reconstitution to vascular regeneration [Miranville et al., 2004; Miyahara et al., 2006; Falanga et al., 2007; Li et al., 2009]. The primary applications of multipotent progenitor cells have been translational cell therapy and tissue engineering, but several groups focused on basic science are also investigating the function and developmental origins of these so-called ‘adult stem’ cells [Crisan et al., 2008; Tang et al., 2008]. They are an attractive option for use in translational technologies because they can be used in autologous approaches to tissue regeneration and replacement without destroying embryonic tissues or introducing pluripotency genes. Although their utility is hindered by a lack of indefinite self-renewal and limited plasticity [Ribitsch et al., 2010], multipotent progenitors have been proven to contribute to certain regenerative metrics when used in tissue regeneration models [Miranville et al., 2004; Falanga et al., 2007; Gordon, 2008]. The sets of lineages toward which a particular multipotent progenitor cell can differentiate are sometimes dependent on the tissue from which the multipotent progenitor cells were isolated [Cavallo et al., 2011; Gaebel et al., 2011; Tallone et al., 2011]. A significant caveat to the use of these cells in a clinical setting is that the success of projects utilizing multipotent progenitor cells is likely to be heavily dependent on the researchers’ ability to deplete irrelevant or counterproductive cell types found in initial isolations while retaining cells possessing therapeutic function [Cavallo et al., 2011; Tallone et al., 2011]. Therefore, the efficient and accurate purification of multipotent progenitor cells is crucial to their utility.
Pluripotent Stem Cells
Pluripotent stem cells have presented vast potential for scientific research since it was discovered that the inner mass cells of an embryo can give rise to all three germ layers of developing mammals. The immediate implication of pluripotency is that inner mass, or ES cells, can be propagated indefinitely in culture and used to study mammalian developmental biology or to develop cell-based regeneration strategies at the organ and tissue levels. While the pluripotency of ES cells offers great potential benefits, it also represents an obstacle to the pursuit of these same goals at the present time. Central to this issue is the exceeding difficulty of directing a pluripotent cell population to a single differentiated cell type without accruing unintended lineages or residual partially undifferentiated subpopulations in the process. The resulting heterogeneous mixture of cell types can sow doubt regarding the validity of experiments and the efficacy and safety of implantable tissues [Kolossov et al., 2006]. Furthermore, it has been well documented that the ethical issues associated with the creation of new ES cell lines, specifically the requisite destruction of an embryo, have slowed their utilization in both the clinic and the laboratory. It is likely that such resistance to ES cell usage will continue for some time.
Fortunately, the less divisive iPS technology has been shown to have many of the same properties of ES cells. iPS cells are created by inducing overexpression of one out of several possible sets of pluripotency genes within somatic cells via one of the genetic insertion methods described above [Takahashi and Yamanaka, 2006; Kim et al., 2008; Nakagawa et al., 2008; Zhang et al., 2009]. The promise of iPS cells in therapeutic applications is such that it may eclipse the usefulness of ES cells, especially when one considers the possibility of reduced immune reaction expected from autologous iPS cells compared to allogeneic ES cell-based strategies. For research into the mechanisms of developmental biology, however, the gold standard is still inner mass-derived ES cells as it has yet to be shown that iPS cells truly retain no vestiges of the differentiated cells from which they are made. Indeed, permanent genetic integration of pluripotency genes when creating iPS cells may be problematic for both therapeutic and basic science applications as it has been reported that even background expression of transgenic pluripotency sequences can cause tumor formation [Okita et al., 2007; Sommer et al., 2010]. Excisable pluripotency cassettes are being used with increasing frequency to create iPS cells; however, the small transgenic sequences remaining after excision are potentially crippling to therapeutic applications [Chang et al., 2009]. New procedures that have been developed for transient expression of pluripotency factors hold great promise for iPS cell usage in clinical applications; however they are technically challenging and induce pluripotent phenotypes at a low rate [Volpers and Kochanek, 2004; Okita et al., 2008; Yu et al., 2009; Jia et al., 2010; Warren et al., 2010].
Immortal Cell Lines
Immortal cells provide researchers with a direct answer to the limited supply of primary cells for experimentation, but they are not functionally relevant to many systems outside of specific applications in cancer research. Most immortalized cells contain a mutation either promoting or inhibiting a gene(s) responsible for the regulation of the cell cycle and proliferation [Rhim, 2000; Gudjonsson et al., 2004]. One of the most common methods for the immortalization of primary cells is the use of the simian vacuolating virus 40 (SV40) viral vector to deliver large T antigen (TAg) or overexpress avian myelocytomatosis virus oncogene cellular homolog (c-Myc) [Ali and DeCaprio, 2001; Gil et al., 2005]. This system is intrinsically valuable in the examination of mutations that influence proliferation, provided the specific means and associated mechanisms of immortalization have been taken into account. Whereas these cells can be very useful when used indirectly in research applications (as they are when used to make viral vectors in vitro), direct usage of immortalized cells in therapeutic applications is problematic as the risks of hyperproliferation and tumor formation are high.
Conclusions
The techniques described above are illustrations of the ingenuity of cell biologists and bioengineers, but they have not solved the problem of cell purification. As science promises and delivers progressively greater milestones, the demand for cures to diseases and FDA-approved therapeutics increases as well. For such achievements to be realized, it is imperative that significant advancements in cell purification methods be made.
As mentioned before, the selection of an appropriate existing cell purification method depends on which cell type will be used, which, in turn, depends on the application for which it will be used. It is, therefore, important to reflect upon the cell source and application when considering the importance of cell purification. It may never be practical to use immortalized cells in therapeutic applications because of their respective drawbacks. Primary cells are the most common cell type used for transplants and grafts, but the supply of human primary cells is too limited for the demands of laboratory research and clinical demand. Multipotent progenitor cells do not have the plasticity for use in major tissue engineering endeavors such as organ fabrication, but they have shown significant regenerative effects in cell therapy applications. Pluripotent stem cells, on the other hand, may not be the most efficient cellular reagent for cell therapy applications; however, they present an opportunity to create entire tissues and organs ex vivo using strategies based on the principles of developmental biology.
Current technologies offer wide ranges of efficiency and mechanisms, but the principals guiding the development of future cell purification methods are relatively constant across all applications. When identifying and separating distinct cell populations, it is important for both basic science research and clinical applications to use techniques that alter cell phenotype as little as possible. This often takes the form of transient, nonintegrating fluorescent markers which are then used in conjunction with FACS to obtain purified populations. It is advantageous to develop high throughput systems capable of processing large quantities of cell mixtures with rare target cell subpopulations. It is also important to maximize efficiency in cell purification methods, meaning that cell loss during sorting should be minimized and the resulting population purity should be high. Although several techniques outlined in this review will undoubtedly continue to play a role in the future of cell purification, it is the innovations to be made that are most interesting.
Acknowledgements
The authors would like to thank Dr. Carolyn Macica of Yale University for providing critical review of the manuscript. This work was supported by the Yale startup fund (E.C.B. and Y.Q.), Connecticut Stem Cell Research Program Grant 09SCAYALE10 (Y.Q.), NIH 1K02HL101990-01 (Y.Q.), UL1 RR024139 (Y.Q.), and NIH 5T32 HL007950 (P.J.A.).
Abbreviations used in this paper
- ANP
atrial natriuretic peptide
- BAC
bacterial artificial chromosome
- c-Myc
avian myelocytomatosis virus oncogene cellular homolog
- cTnI
cardiac troponin I
- cTnT
cardiac troponin T
- EMILIN2
elastin microfibril interface 2
- ES
embryonic stem
- FACS
fluorescence-activated cell sorting
- GATA4
GATA transcription factor 4
- GFP
green fluorescent protein
- HCN4
hyperpolarization-activated cyclic nucleotide-gated potassium channel 4
- HUVECs
human umbilical vein endothelial cells
- iPS
induced pluripotent stem
- mlc2a
myosin light chain 2a
- mlc2v
myosin light chain 2v
- Nkx2.5
NK2 transcription-related factor locus 5
- SV40
simian vacuolating virus 40
- TAg
large T antigen
- TMRM
tetramethylrhodamine methyl
References
- Adam Young, D., J.A. Dequach, K.L. Christman (2010) Human cardiomyogenesis and the need for systems biology analysis. Wiley Interdiscip Rev Syst Biol Med, E-pub ahead of print. [DOI] [PMC free article] [PubMed]
- Adams A.A., Okagbare P.I., Feng J., Hupert M.L., Patterson D., Gottert J., McCarley R.L., Nikitopoulos D., Murphy M.C., Soper S.A. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J Am Chem Soc. 2008;130:8633–8641. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S.H., DeCaprio J.A. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin Cancer Biol. 2001;11:15–23. doi: 10.1006/scbi.2000.0342. [DOI] [PubMed] [Google Scholar]
- Arcidiacono J.A., Evdokimov E., Lee M.H., Jones J., Rudenko L., Schneider B., Snoy P.J., Wei C.H., Wensky A.K., Wonnacott K. Regulation of xenogeneic porcine pancreatic islets. Xenotransplantation. 2010;17:329–337. doi: 10.1111/j.1399-3089.2010.00592.x. [DOI] [PubMed] [Google Scholar]
- Batista Lobo S., Denyer M., Britland S., Javid F.A. Development of an intestinal cell culture model to obtain smooth muscle cells and myenteric neurones. J Anat. 2007;211:819–829. doi: 10.1111/j.1469-7580.2007.00820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudino T.A., Carver W., Giles W., Borg T.K. Cardiac fibroblasts: friend or foe? Am J Physiol Heart Circ Physiol. 2006;291:H1015–H1026. doi: 10.1152/ajpheart.00023.2006. [DOI] [PubMed] [Google Scholar]
- Blackman B.R., Garcia-Cardena G., Gimbrone M.A., Jr A new in vitro model to evaluate differential responses of endothelial cells to simulated arterial shear stress waveforms. J Biomech Eng. 2002;124:397–407. doi: 10.1115/1.1486468. [DOI] [PubMed] [Google Scholar]
- Blin G., Nury D., Stefanovic S., Neri T., Guillevic O., Brinon B., Bellamy V., Rucker-Martin C., Barbry P., Bel A., Bruneval P., Cowan C., Pouly J., Mitalipov S., Gouadon E., Binder P., Hagege A., Desnos M., Renaud J.F., Menasche P., Puceat M. A purified population of multipotent cardiovascular progenitors derived from primate pluripotent stem cells engrafts in postmyocardial infarcted nonhuman primates. J Clin Invest. 2010;120:1125–1139. doi: 10.1172/JCI40120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobis-Wozowicz S., Osiak A., Rahman S.H., Cathomen T. Targeted genome editing in pluripotent stem cells using zinc-finger nucleases. Methods. 2011;53:339–346. doi: 10.1016/j.ymeth.2010.12.019. [DOI] [PubMed] [Google Scholar]
- Bu L., Jiang X., Martin-Puig S., Caron L., Zhu S., Shao Y., Roberts D.J., Huang P.L., Domian I.J., Chien K.R. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- Cavallo C., Cuomo C., Fantini S., Ricci F., Tazzari P.L., Lucarelli E., Donati D., Facchini A., Lisignoli G., Fornasari P.M., Grigolo B., Moroni L. Comparison of alternative mesenchymal stem cell sources for cell banking and musculoskeletal advanced therapies. J Cell Biochem. 2011;112:1418–1430. doi: 10.1002/jcb.23058. [DOI] [PubMed] [Google Scholar]
- Chalberg T.W., Portlock J.L., Olivares E.C., Thyagarajan B., Kirby P.J., Hillman R.T., Hoelters J., Calos M.P. Integration specificity of phage phiC31 integrase in the human genome. J Mol Biol. 2006;357:28–48. doi: 10.1016/j.jmb.2005.11.098. [DOI] [PubMed] [Google Scholar]
- Chang C.W., Lai Y.S., Pawlik K.M., Liu K., Sun C.W., Li C., Schoeb T.R., Townes T.M. Polycistronic lentiviral vector for ‘hit and run’ reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:1042–1049. doi: 10.1002/stem.39. [DOI] [PubMed] [Google Scholar]
- Chen J., Kubalak S.W., Minamisawa S., Price R.L., Becker K.D., Hickey R., Ross J., Jr, Chien K.R. Selective requirement of myosin light chain 2v in embryonic heart function. J Biol Chem. 1998;273:1252–1256. doi: 10.1074/jbc.273.2.1252. [DOI] [PubMed] [Google Scholar]
- Crisan M., Yap S., Casteilla L., Chen C.W., Corselli M., Park T.S., Andriolo G., Sun B., Zheng B., Zhang L., Norotte C., Teng P.N., Traas J., Schugar R., Deasy B.M., Badylak S., Buhring H.J., Giacobino J.P., Lazzari L., Huard J., Peault B. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Cui X., Ji D., Fisher D.A., Wu Y., Briner D.M., Weinstein E.J. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol. 2011;29:64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- Davis R.P., Costa M., Grandela C., Holland A.M., Hatzistavrou T., Micallef S.J., Li X., Goulburn A.L., Azzola L., Elefanty A.G., Stanley E.G. A protocol for removal of antibiotic resistance cassettes from human embryonic stem cells genetically modified by homologous recombination or transgenesis. Nat Protoc. 2008;3:1550–1558. doi: 10.1038/nprot.2008.146. [DOI] [PubMed] [Google Scholar]
- Davison J.M., Woo Park S., Rhee J.M., Leach S.D. Characterization of Kras-mediated pancreatic tumorigenesis in zebrafish. Methods Enzymol. 2008;438:391–417. doi: 10.1016/S0076-6879(07)38027-0. [DOI] [PubMed] [Google Scholar]
- Diaz-Prado, S., E. Muinos-Lopez, T. Hermida-Gomez, M.E. Rendal-Vazquez, I. Fuentes-Boquete, F.J. de Toro, F.J. Blanco (2010) Isolation and characterization of mesenchymal stem cells from human amniotic membrane. Tissue Eng Part C Methods, E-pub ahead of print. [DOI] [PubMed]
- Dittel B.N. Depletion of specific cell populations by complement depletion. J Vis Exp. 2010;5:1487. doi: 10.3791/1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrzynski H., Nikolski V.P., Sambelashvili A.T., Greener I.D., Yamamoto M., Boyett M.R., Efimov I.R. Site of origin and molecular substrate of atrioventricular junctional rhythm in the rabbit heart. Circ Res. 2003;93:1102–1110. doi: 10.1161/01.RES.0000101913.95604.B9. [DOI] [PubMed] [Google Scholar]
- Ebbe M. The 2000 Global Laboratory Product Usage Study. San Carlos: PhorTech International; 2000. [Google Scholar]
- Falanga V., Iwamoto S., Chartier M., Yufit T., Butmarc J., Kouttab N., Shrayer D., Carson P. Autologous bone marrow-derived cultured mesenchymal stem cells delivered in a fibrin spray accelerate healing in murine and human cutaneous wounds. Tissue Eng. 2007;13:1299–1312. doi: 10.1089/ten.2006.0278. [DOI] [PubMed] [Google Scholar]
- Freund D., Fonseca A.V., Janich P., Bornhauser M., Corbeil D. Differential expression of biofunctional GM1 and GM3 gangliosides within the plastic-adherent multipotent mesenchymal stromal cell population. Cytotherapy. 2010;12:131–142. doi: 10.3109/14653240903476438. [DOI] [PubMed] [Google Scholar]
- Fu J.D., Jiang P., Rushing S., Liu J., Chiamvimonvat N., Li R.A. Na+/Ca2+ exchanger is a determinant of excitation-contraction coupling in human embryonic stem cell-derived ventricular cardiomyocytes. Stem Cells Dev. 2010;19:773–782. doi: 10.1089/scd.2009.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebel R., Furlani D., Sorg H., Polchow B., Frank J., Bieback K., Wang W., Klopsch C., Ong L.L., Li W., Ma N., Steinhoff G. Cell origin of human mesenchymal stem cells determines a different healing performance in cardiac regeneration. PLoS One. 2011;6:e15652. doi: 10.1371/journal.pone.0015652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil J., Kerai P., Lleonart M., Bernard D., Cigudosa J.C., Peters G., Carnero A., Beach D. Immortalization of primary human prostate epithelial cells by c-Myc. Cancer Res. 2005;65:2179–2185. doi: 10.1158/0008-5472.CAN-03-4030. [DOI] [PubMed] [Google Scholar]
- Gordon M.Y. Stem cells for regenerative medicine – biological attributes and clinical application. Exp Hematol. 2008;36:726–732. doi: 10.1016/j.exphem.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Gudjonsson T., Villadsen R., Ronnov-Jessen L., Petersen O.W. Immortalization protocols used in cell culture models of human breast morphogenesis. Cell Mol Life Sci. 2004;61:2523–2534. doi: 10.1007/s00018-004-4167-z. [DOI] [PubMed] [Google Scholar]
- Harris J.E. Reprogramming the future of stem cell patents. Ann Health Law Adv Directive. 2008;18:10–15. [Google Scholar]
- Hattori F., Chen H., Yamashita H., Tohyama S., Satoh Y.S., Yuasa S., Li W., Yamakawa H., Tanaka T., Onitsuka T., Shimoji K., Ohno Y., Egashira T., Kaneda R., Murata M., Hidaka K., Morisaki T., Sasaki E., Suzuki T., Sano M., Makino S., Oikawa S., Fukuda K. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat Methods. 2010;7:61–66. doi: 10.1038/nmeth.1403. [DOI] [PubMed] [Google Scholar]
- Hempel C.M., Sugino K., Nelson S.B. A manual method for the purification of fluorescently labeled neurons from the mammalian brain. Nat Protoc. 2007;2:2924–2929. doi: 10.1038/nprot.2007.416. [DOI] [PubMed] [Google Scholar]
- Hermening S., Kugler S., Bahr M., Isenmann S. Improved high-capacity adenoviral vectors for high-level neuron-restricted gene transfer to the CNS. J Virol Methods. 2006;136:30–37. doi: 10.1016/j.jviromet.2006.03.031. [DOI] [PubMed] [Google Scholar]
- Hidaka K., Shirai M., Lee J.K., Wakayama T., Kodama I., Schneider M.D., Morisaki T. The cellular prion protein identifies bipotential cardiomyogenic progenitors. Circ Res. 2010;106:111–119. doi: 10.1161/CIRCRESAHA.109.209478. [DOI] [PubMed] [Google Scholar]
- Horn P., Bork S., Diehlmann A., Walenda T., Eckstein V., Ho A.D., Wagner W. Isolation of human mesenchymal stromal cells is more efficient by red blood cell lysis. Cytotherapy. 2008;10:676–685. doi: 10.1080/14653240802398845. [DOI] [PubMed] [Google Scholar]
- Huang T. Stem cell patent landscape and patent strategy. Trends Bio/Pharm Ind. 2010;6:41–46. [Google Scholar]
- Huber I., Itzhaki I., Caspi O., Arbel G., Tzukerman M., Gepstein A., Habib M., Yankelson L., Kehat I., Gepstein L. Identification and selection of cardiomyocytes during human embryonic stem cell differentiation. FASEB J. 2007;21:2551–2563. doi: 10.1096/fj.05-5711com. [DOI] [PubMed] [Google Scholar]
- Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F., Wilson K.D., Sun N., Gupta D.M., Huang M., Li Z., Panetta N.J., Chen Z.Y., Robbins R.C., Kay M.A., Longaker M.T., Wu J.C. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing D., Fonseca A.V., Alakel N., Fierro F.A., Muller K., Bornhauser M., Ehninger G., Corbeil D., Ordemann R. Hematopoietic stem cells in co-culture with mesenchymal stromal cells – modeling the niche compartments in vitro. Haematologica. 2010;95:542–550. doi: 10.3324/haematol.2009.010736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers J., Berns A. Conditional mouse models of sporadic cancer. Nat Rev Cancer. 2002;2:251–265. doi: 10.1038/nrc777. [DOI] [PubMed] [Google Scholar]
- Kalpana G.V. Retroviral vectors for liver-directed gene therapy. Semin Liver Dis. 1999;19:27–37. doi: 10.1055/s-2007-1007095. [DOI] [PubMed] [Google Scholar]
- Kim J.B., Zaehres H., Wu G., Gentile L., Ko K., Sebastiano V., Arauzo-Bravo M.J., Ruau D., Han D.W., Zenke M., Scholer H.R. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- Klimatcheva E., Rosenblatt J.D., Planelles V. Lentiviral vectors and gene therapy. Front Biosci. 1999;4:D481–D496. doi: 10.2741/klimatcheva. [DOI] [PubMed] [Google Scholar]
- Koller M.R., Hanania E.G., Stevens J., Eisfeld T.M., Sasaki G.C., Fieck A., Palsson B.O. High-throughput laser-mediated in situ cell purification with high purity and yield. Cytometry A. 2004;61:153–161. doi: 10.1002/cyto.a.20079. [DOI] [PubMed] [Google Scholar]
- Kolossov E., Bostani T., Roell W., Breitbach M., Pillekamp F., Nygren J.M., Sasse P., Rubenchik O., Fries J.W., Wenzel D., Geisen C., Xia Y., Lu Z., Duan Y., Kettenhofen R., Jovinge S., Bloch W., Bohlen H., Welz A., Hescheler J., Jacobsen S.E., Fleischmann B.K. Engraftment of engineered ES cell-derived cardiomyocytes but not BM cells restores contractile function to the infarcted myocardium. J Exp Med. 2006;203:2315–2327. doi: 10.1084/jem.20061469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubalak S.W., Miller-Hance W.C., O'Brien T.X., Dyson E., Chien K.R. Chamber specification of atrial myosin light chain-2 expression precedes septation during murine cardiogenesis. J Biol Chem. 1994;269:16961–16970. [PubMed] [Google Scholar]
- Kumar P., Woon-Khiong C. Optimization of lentiviral vectors generation for biomedical and clinical research purposes: contemporary trends in technology development and applications. Curr Gene Ther. 2011;11:144–153. doi: 10.2174/156652311794940782. [DOI] [PubMed] [Google Scholar]
- Laflamme M.A., Chen K.Y., Naumova A.V., Muskheli V., Fugate J.A., Dupras S.K., Reinecke H., Xu C., Hassanipour M., Police S., O'Sullivan C., Collins L., Chen Y., Minami E., Gill E.A., Ueno S., Yuan C., Gold J., Murry C.E. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- Lew A.M., Glogauer M., McUlloch C.A. Specific inhibition of skeletal alpha-actin gene transcription by applied mechanical forces through integrins and actin. Biochem J. 1999;341:647–653. [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chen S., Yuan J., Yang Y., Li J., Ma J., Wu X., Freund M., Pollok K., Hanenberg H., Goebel W.S., Yang F.C. Mesenchymal stem/progenitor cells promote the reconstitution of exogenous hematopoietic stem cells in Fancg–/– mice in vivo. Blood. 2009;113:2342–2351. doi: 10.1182/blood-2008-07-168138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang L., Fatahi R., Kronenberg M., Kalajzic I., Rowe D., Li Y., Maye P. Isolation of murine bone marrow derived mesenchymal stem cells using Twist2 Cre transgenic mice. Bone. 2010;47:916–925. doi: 10.1016/j.bone.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Fan T., Zhao Q., Zeng W., Zaslavsky E., Chen J.J., Frohman M.A., Golightly M.G., Madajewicz S., Chen W.T. Isolation of circulating epithelial and tumor progenitor cells with an invasive phenotype from breast cancer patients. Int J Cancer. 2010;126:669–683. doi: 10.1002/ijc.24814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. Recombinant adeno-associated virus as delivery vector for gene therapy – a review. Stem Cells Dev. 2004;13:133–145. doi: 10.1089/154732804773099335. [DOI] [PubMed] [Google Scholar]
- Martin-Puig S., Wang Z., Chien K.R. Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell. 2008;2:320–331. doi: 10.1016/j.stem.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Mejia-Pous C., Vinuelas J., Faure C., Koszela J., Kawakami K., Takahashi Y., Gandrillon O. A combination of transposable elements and magnetic cell sorting provides a very efficient transgenesis system for chicken primary erythroid progenitors. BMC Biotechnol. 2009;9:81. doi: 10.1186/1472-6750-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metezeau P. Image and flow cytometry: companion techniques for adherent and non-adherent cell analysis and sorting. Biol Cell. 1993;78:129–134. doi: 10.1016/0248-4900(93)90123-v. [DOI] [PubMed] [Google Scholar]
- Miranville A., Heeschen C., Sengenes C., Curat C.A., Busse R., Bouloumie A. Improvement of postnatal neovascularization by human adipose tissue-derived stem cells. Circulation. 2004;110:349–355. doi: 10.1161/01.CIR.0000135466.16823.D0. [DOI] [PubMed] [Google Scholar]
- Miyahara Y., Nagaya N., Kataoka M., Yanagawa B., Tanaka K., Hao H., Ishino K., Ishida H., Shimizu T., Kangawa K., Sano S., Okano T., Kitamura S., Mori H. Monolayered mesenchymal stem cells repair scarred myocardium after myocardial infarction. Nat Med. 2006;12:459–465. doi: 10.1038/nm1391. [DOI] [PubMed] [Google Scholar]
- Moravcikova J., Vaculkova E., Bauer M., Libantova J. Feasibility of the seed specific cruciferin C promoter in the self excision Cre/loxP strategy focused on generation of marker-free transgenic plants. Theor Appl Genet. 2008;117:1325–1334. doi: 10.1007/s00122-008-0866-4. [DOI] [PubMed] [Google Scholar]
- Muller M., Fleischmann B.K., Selbert S., Ji G.J., Endl E., Middeler G., Muller O.J., Schlenke P., Frese S., Wobus A.M., Hescheler J., Katus H.A., Franz W.M. Selection of ventricular-like cardiomyocytes from ES cells in vitro. FASEB J. 2000;14:2540–2548. doi: 10.1096/fj.00-0002com. [DOI] [PubMed] [Google Scholar]
- Mummery C., Ward-van Oostwaard D., Doevendans P., Spijker R., van den Brink S., Hassink R., van der Heyden M., Opthof T., Pera M., de la Riviere A.B., Passier R., Tertoolen L. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107:2733–2740. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Nelson T.J., Martinez-Fernandez A., Terzic A. Induced pluripotent stem cells: developmental biology to regenerative medicine. Nat Rev Cardiol. 2010;7:700–710. doi: 10.1038/nrcardio.2010.159. [DOI] [PubMed] [Google Scholar]
- Nemir M., Croquelois A., Pedrazzini T., Radtke F. Induction of cardiogenesis in embryonic stem cells via downregulation of Notch1 signaling. Circ Res. 2006;98:1471–1478. doi: 10.1161/01.RES.0000226497.52052.2a. [DOI] [PubMed] [Google Scholar]
- Nohe B., Zanke C., Johannes T., Kiefer T., Dieterich H.J. Effects of magnetic cell separation on monocyte adhesion to endothelial cells under flow. APMIS. 2002;110:299–308. doi: 10.1034/j.1600-0463.2002.100404.x. [DOI] [PubMed] [Google Scholar]
- Nomura T., Ashihara E., Tateishi K., Ueyama T., Takahas-Hi T., Yamagishi M., Kubo T., Yaku H., Matsubara H., Oh H. Therapeutic potential of stem/progenitor cells in human skeletal muscle for cardiovascular regeneration. Curr Stem Cell Res Ther. 2007;2:293–300. doi: 10.2174/157488807782793808. [DOI] [PubMed] [Google Scholar]
- O'Connell T.D., Rodrigo M.C., Simpson P.C. Isolation and culture of adult mouse cardiac myocytes. Methods Mol Biol. 2007;357:271–296. doi: 10.1385/1-59745-214-9:271. [DOI] [PubMed] [Google Scholar]
- Ohnuma K., Yomo T., Asashima M., Kaneko K. Sorting of cells of the same size, shape, and cell cycle stage for a single cell level assay without staining. BMC Cell Biol. 2006;7:25. doi: 10.1186/1471-2121-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Okita K., Nakagawa M., Hyenjong H., Ichisaka T., Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Pamukcu B., Lip G.Y., Devitt A., Griffiths H., Shantsila E. The role of monocytes in atherosclerotic coronary artery disease. Ann Med. 2010;42:394–403. doi: 10.3109/07853890.2010.497767. [DOI] [PubMed] [Google Scholar]
- Philpott N.J., Thrasher A.J. Use of nonintegrating lentiviral vectors for gene therapy. Hum Gene Ther. 2007;18:483–489. doi: 10.1089/hum.2007.013. [DOI] [PubMed] [Google Scholar]
- Rhim J.S. Development of human cell lines from multiple organs. Ann NY Acad Sci. 2000;919:16–25. doi: 10.1111/j.1749-6632.2000.tb06863.x. [DOI] [PubMed] [Google Scholar]
- Ribitsch I., Burk J., Delling U., Geissler C., Gittel C., Julke H., Brehm W. Basic science and clinical application of stem cells in veterinary medicine. Adv Biochem Eng Biotechnol. 2010;123:219–263. doi: 10.1007/10_2010_66. [DOI] [PubMed] [Google Scholar]
- Sarov M., Schneider S., Pozniakovski A., Roguev A., Ernst S., Zhang Y., Hyman A.A., Stewart A.F. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat Methods. 2006;3:839–844. doi: 10.1038/nmeth933. [DOI] [PubMed] [Google Scholar]
- Schauwer C.D., Meyer E., Walle G.R., Soom A.V. Markers of stemness in equine mesenchymal stem cells: a plea for uniformity. Theriogenology. 2011;75:1431–1443. doi: 10.1016/j.theriogenology.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Schindler M., Allen M.L., Olinger M.R., Holland J.F. Automated analysis and survival selection of anchorage-dependent cells under normal growth conditions. Cytometry. 1985;6:368–374. doi: 10.1002/cyto.990060415. [DOI] [PubMed] [Google Scholar]
- Shibano T., Takeda M., Suetake I., Kawakami K., Asashima M., Tajima S., Taira M. Recombinant Tol2 transposase with activity in Xenopus embryos. FEBS Lett. 2007;581:4333–4336. doi: 10.1016/j.febslet.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Simek S.L. Characterization of gene therapy products and the impact of manufacturing changes on product comparability. Dev Biol (Basel) 2005;122:139–144. [PubMed] [Google Scholar]
- Sommer C.A., Sommer A.G., Longmire T.A., Christodoulou C., Thomas D.D., Gostissa M., Alt F.W., Murphy G.J., Kotton D.N., Mostoslavsky G. Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector. Stem Cells. 2010;28:64–74. doi: 10.1002/stem.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Chung S.K., Xu Y. Modeling disease in human ESCs using an efficient BAC-based homologous recombination system. Cell Stem Cell. 2010;6:80–89. doi: 10.1016/j.stem.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Szaniszlo P., Rose W.A., Wang N., Reece L.M., Tsulaia T.V., Hanania E.G., Elferink C.J., Leary J.F. Scanning cytometry with a LEAP: laser-enabled analysis and processing of live cells in situ. Cytometry A. 2006;69:641–651. doi: 10.1002/cyto.a.20291. [DOI] [PubMed] [Google Scholar]
- Szaniszlo P., Wang N., Sinha M., Reece L.M., Van Hook J.W., Luxon B.A., Leary J.F. Getting the right cells to the array: gene expression microarray analysis of cell mixtures and sorted cells. Cytometry A. 2004;59:191–202. doi: 10.1002/cyto.a.20055. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tallone T., Realini C., Bohmler A., Kornfeld C., Vassalli G., Moccetti T., Bardelli S., Soldati G. Adult human adipose tissue contains several types of multipotent cells. J Cardiovasc Transl Res. 2011;4:200–210. doi: 10.1007/s12265-011-9257-3. [DOI] [PubMed] [Google Scholar]
- Tang W., Zeve D., Suh J.M., Bosnakovski D., Kyba M., Hammer R.E., Tallquist M.D., Graff J.M. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzen T., Zembowicz F., Cowan C.A. Genome modification in human embryonic stem cells. J Cell Physiol. 2010;222:278–281. doi: 10.1002/jcp.21948. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B., Liu Y., Shin S., Lakshmipathy U., Scheyhing K., Xue H., Ellerstrom C., Strehl R., Hyllner J., Rao M.S., Chesnut J.D. Creation of engineered human embryonic stem cell lines using phiC31 integrase. Stem Cells. 2008;26:119–126. doi: 10.1634/stemcells.2007-0283. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B., Olivares E.C., Hollis R.P., Ginsburg D.S., Calos M.P. Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Mol Cell Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., James S., Zuo J., Fritzsch B., Beisel K.W. Conditional and inducible gene recombineering in the mouse inner ear. Brain Res. 2006;1091:243–254. doi: 10.1016/j.brainres.2006.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof D., Dormeyer W., Braam S.R., Passier R., Monshouwer-Kloots J., Ward-van Oostwaard D., Heck A.J., Krijgsveld J., Mummery C.L. Identification of cell surface proteins for antibody-based selection of human embryonic stem cell-derived cardiomyocytes. J Proteome Res. 2010;9:1610–1618. doi: 10.1021/pr901138a. [DOI] [PubMed] [Google Scholar]
- Volpers C., Kochanek S. Adenoviral vectors for gene transfer and therapy. J Gene Med. 2004;6(suppl 1):S164–S171. doi: 10.1002/jgm.496. [DOI] [PubMed] [Google Scholar]
- Vondran F.W., Katenz E., Schwartlander R., Morgul M.H., Raschzok N., Gong X., Cheng X., Kehr D., Sauer I.M. Isolation of primary human hepatocytes after partial hepatectomy: criteria for identification of the most promising liver specimen. Artif Organs. 2008;32:205–213. doi: 10.1111/j.1525-1594.2007.00524.x. [DOI] [PubMed] [Google Scholar]
- Warren L., Manos P.D., Ahfeldt T., Loh Y.H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A., Daley G.Q., Brack A.S., Collins J.J., Cowan C., Schlaeger T.M., Rossi D.J. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A.P., Tata J.R. Primary culture, cellular stress and differentiated function. FEBS Lett. 1984;176:8–15. doi: 10.1016/0014-5793(84)80902-3. [DOI] [PubMed] [Google Scholar]
- Wu S.C., Meir Y.J., Coates C.J., Handler A.M., Pelczar P., Moisyadi S., Kaminski J.M. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc Natl Acad Sci USA. 2006;103:15008–15013. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Police S., Hassanipour M., Gold J.D. Cardiac bodies: a novel culture method for enrichment of cardiomyocytes derived from human embryonic stem cells. Stem Cells Dev. 2006;15:631–639. doi: 10.1089/scd.2006.15.631. [DOI] [PubMed] [Google Scholar]
- Yang L., Soonpaa M.H., Adler E.D., Roepke T.K., Kattman S.J., Kennedy M., Henckaerts E., Bonham K., Abbott G.W., Linden R.M., Field L.J., Keller G.M. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Yona, S., R. Hayhoe, I. Avraham-Davidi (2010) Monocyte and neutrophil isolation and migration assays. Curr Protoc Immunol chapter 14: unit 14.15. [DOI] [PubMed]
- Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I.I., Thomson J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa S., Itabashi Y., Koshimizu U., Tanaka T., Sugimura K., Kinoshita M., Hattori F., Fukami S., Shimazaki T., Ogawa S., Okano H., Fukuda K. Transient inhibition of BMP signaling by Noggin induces cardiomyocyte differentiation of mouse embryonic stem cells. Nat Biotechnol. 2005;23:607–611. doi: 10.1038/nbt1093. [DOI] [PubMed] [Google Scholar]
- Zborowski M., Moore L.R., Reddy S., Chen G.H., Sun L., Chalmers J.J. Magnetic flow sorting using a model system of human lymphocytes and a colloidal magnetic label. ASAIO J. 1996;42:M666–671. doi: 10.1097/00002480-199609000-00071. [DOI] [PubMed] [Google Scholar]
- Zhang J., Wilson G.F., Soerens A.G., Koonce C.H., Yu J., Palecek S.P., Thomson J.A., Kamp T.J. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104:e30–41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Hirai M., Cantero S., Ciubotariu R., Dobrila L., Hirsh A., Igura K., Satoh H., Yokomi I., Nishimura T., Yamaguchi S., Yoshimura K., Rubinstein P., Takahashi T.A. Isolation and characterization of mesenchymal stem cells from human umbilical cord blood: reevaluation of critical factors for successful isolation and high ability to proliferate and differentiate to chondrocytes as compared to mesenchymal stem cells from bone marrow and adipose tissue. J Cell Biochem. 2011;112:1206–1218. doi: 10.1002/jcb.23042. [DOI] [PubMed] [Google Scholar]
- Zuidema M.Y., Zhang C. Ischemia/reperfusion injury: the role of immune cells. World J Cardiol. 2010;2:325–332. doi: 10.4330/wjc.v2.i10.325. [DOI] [PMC free article] [PubMed] [Google Scholar]