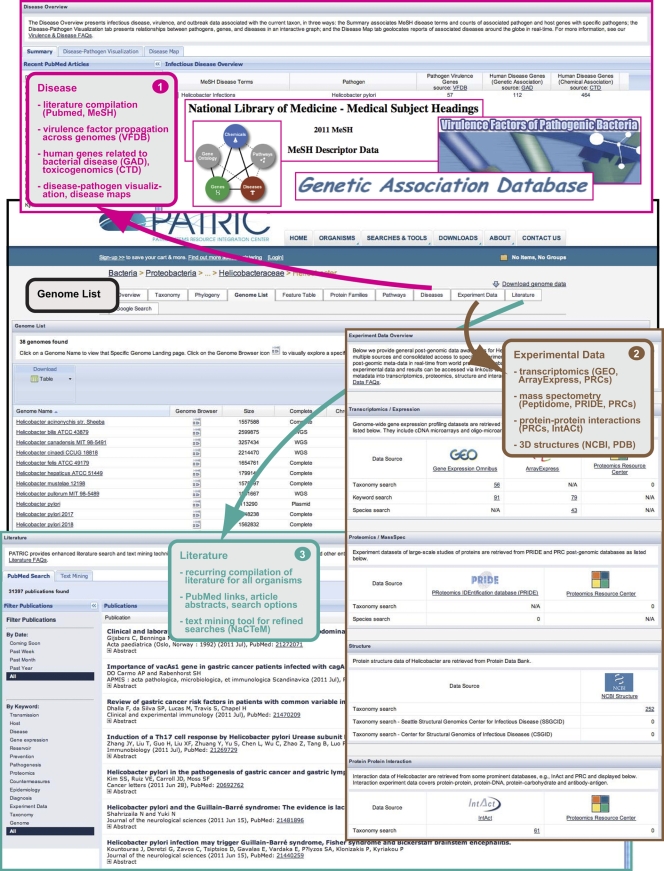
Fig. 4.
Schema depicting the integrated community-derived associated data available from an organism “Overview” homepage. Navigation from the Helicobacter “Genome List” (outlined in black) is illustrated. Disease information (box 1) can be summarized into four main categories: Literature (PubMed article compilation and MeSH terms for database searching [32]), virulence factors (data from the Virulence Factor Database [VFDB] [54] is used to identify all putative homologs present within other bacterial genomes), human genes associated with disease (Genetic Association Database [8, 57] and Comparative Toxicogenomics Database [14]), and disease-pathogen data (interactive graphics for relationships between pathogens, diseases, virulence genes, and disease-associated host genes, as well as interactive global health maps [11] illustrating recent reports and outbreaks of bacterial diseases). “Experimental Data” (box 2) encompasses transcriptomic data (GEO [6, 7], ArrayExpress [26], and Proteomics Resource Centers [PRCs] [56]), proteomics data from mass spectrometry (Peptidome [25], PRIDE [48] and the PRCs), protein-protein interaction data from the PRCs and IntACt (4), and protein 3-D structure data from NCBI and Protein Data Bank (PDB) (10). “Literature” (box 3) is primarily comprised of a recurrent compilation of literature and web text resources pertaining to each organism (PubMed abstracts and links to articles), with a search tool that allows filtering by keywords, dates, etc. An integrated text-mining tool (UK National Text Mining Centre [NaCTeM]) allows efficient recall of relevant documents through the identification of key entities from the search text (i.e., genes, proteins, metabolites, drugs, diseases, symptoms, etc.).