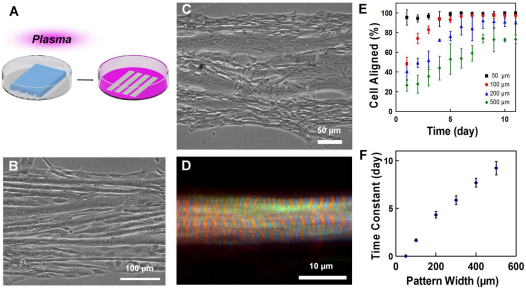
Fig. 1.
Self-organization of myoblasts and geometric alignment of myotubes on plasma patterned substrates. (A) Plasma lithography to create chemical patterns for guiding cell alignment. Areas exposed to plasma (pink) present surface functional groups that facilitate cell adhesion, whereas areas shielded by PDMS (blue structure) prevent cell adhesion. (B) C2C12 mouse myoblasts, and (C) primary chick myoblasts guided on line patterns form linear, aligned myotubes parallel to the boundaries. (D) Sarcomeres in patterned primary chicken skeletal muscle fibers. F-actin is labeled by phalloidin (green), Z-discs by α-actinin (blue) and M-line by titin T114 (red). (E) The temporal evolution of myoblasts aligned to the line patterns with different widths. (F) Time constants of myoblast alignment on different pattern widths. Error bars in E and F are s.d. and s.e. of the mean, respectively.