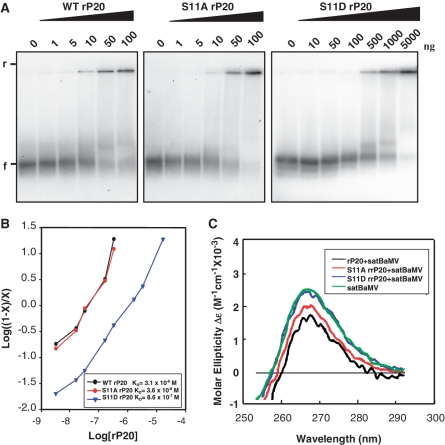
Figure 3.
RNA binding activity of wild-type and mutant rP20. (A) EMSA. Indicated amounts of WT, S11A or S11D rP20 were co-incubated with 5 ng 32P-labeled WT satBaMV transcripts and resolved on 1% non-denaturing agarose gel electrophoresis in 0.5 X TBE buffer (40 mM Tris-boric acid, 1 mM EDTA, pH 8.0) at 150 V and 4°C for 1 h. The gel was analyzed by PhosphorImager. ‘f’ and ‘r’ indicate free and retarded satBaMV RNA, respectively. (B) EMSA data underwent Hill transformation by ImageQuant v.5.2. Data are plotted by log [(1 − x)/x] versus log rP20 concentration, where x is [RNA]/[RNA]total). The slope of best-fit equation determines the Hill coefficient. The dissociation constants (KD) for WT, S11A and S11D rP20 are 3.1 × 10−8 M, 3.6 × 10−8 M and 8.6 × 10−7 M, respectively. (C) Circular dichroism (CD) spectra of satBaMV RNA. In total, 500 ng of WT satBaMV transcripts were incubated with 50 ng WT, S11A or S11D rP20 in PBS buffer (50 mM sodium phosphate, pH 7.0, 150 mM NaCl) on ice for 30 min. SatBaMV transcripts not pre-incubated with rP20 were included as a control. RNA ellipticity was recorded in near-UV region (250–300 nm), and CD is expressed in molar ellipticity.