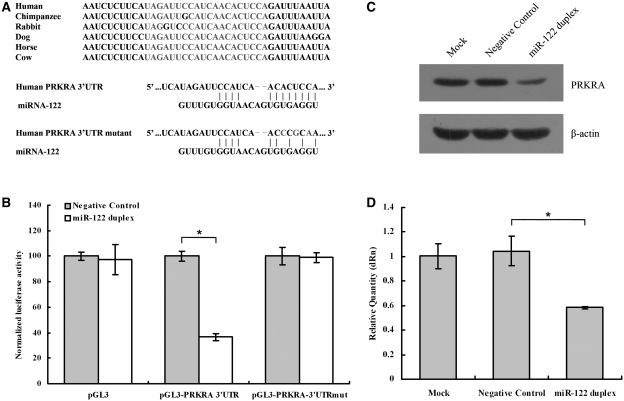
Figure 3.
miR-122 inhibits PRKRA expression. (A upper panel) The target site of miR-122 in PRKRA 3′-UTR is conserved among mammalian species. (A lower panel) Predicted duplex formation between miR-122 and the targeted PRKRA 3′-UTR. The PRKRA 3′-UTR mutant is identical with the wild-type except that its three point substitutions disrupting pairing to miR-122 seed. (B) pGL3-PRKRA-3′-UTR reporter plasmid in which the luciferase-coding sequence had been fused to the 3′-UTR of PRKRA was cotransfected into HepG2 cells with Negative Control (grey columns) or miR-122 duplex (white columns). Luciferase activity was normalized relative to a simultaneously transfected Renilla expression plasmid. 3′-UTR-Mut indicates the introduction of alterations into the seed complementary sites shown in Figure 3A. Results of the mean of quadruplicate assays with standard deviation of the mean are presented. *P < 0.05. (C) Western blots of PRKRA from HepG2 cells. Cells were transfected with Negative Control or miR-122 duplex. Cells were harvested 48 h later, and 30 μg of whole-cell lysate was added into each lane. A β-actin antibody was used in a reprobing as a loading control. (D) Real-time RT–PCR of PRKRA in HepG2 cells transfected with the Negative Control or miR-122 duplex for 48 h. Data were normalized to the level of GAPDH mRNA. Results of the mean of triplicate qPCR assays with standard deviation of the mean are presented. *P < 0.05.