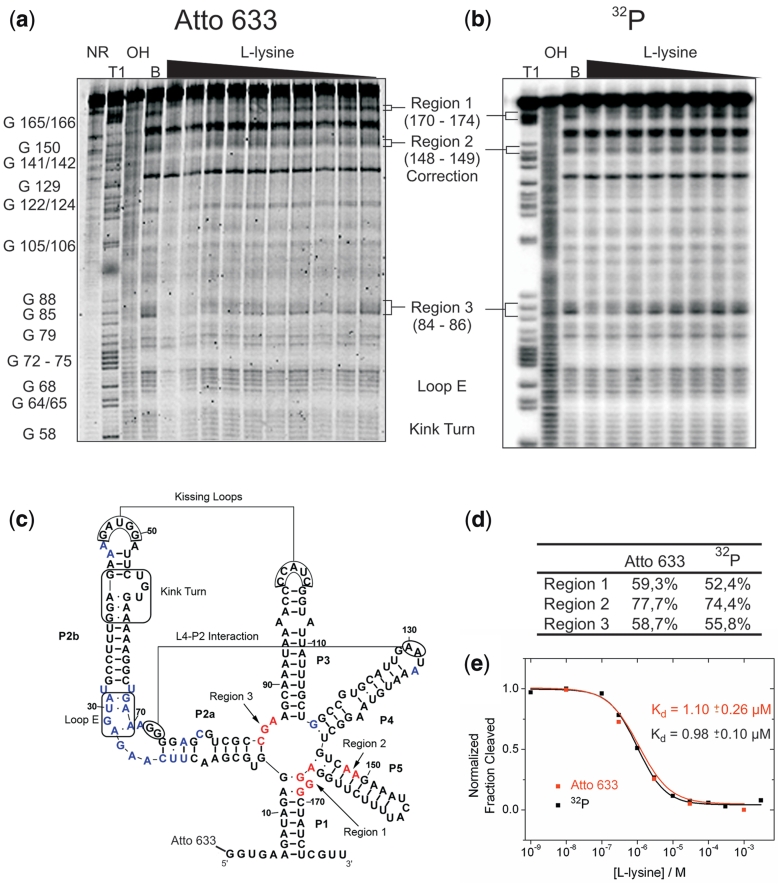
Figure 1.
A fluorescent label can replace a 32P in riboswitch binding studies. (a) In-line probing analysis of the 5′-Atto 633 labelled lysC lysine riboswitch aptamer from B. subtilis. Reactions were performed in the absence (lane B) or in the presence of l-lysine (1 nM to 1 mM). NR, T1 and OH represent unreacted RNA, a G-specific sequencing ladder and a partial alkaline digest, respectively. Selected bands in the T1 lane are annotated with the nucleotide position. Regions 1–3 represent sites where the extent of spontaneous cleavage is modulated by the lysine concentration and were assigned either directly or by analogy to published probing patterns (32,34). (b) In-line probing analysis of the 5′-32P-labelled lysC lysine riboswitch aptamer. (c) Secondary structure representing the lysine riboswitch aptamer. Important tertiary interactions and structural elements are marked. Nucleotides depicted in blue show spontaneous cleavage under in-line probing conditions. Nucleotides depicted in red show lysine-induced changes of their spontaneous cleavage. (d) Comparison of the intensity of the cleavage bands in all three regions of lysine-induced cleavage modulation. For both labels, the intensity of the cleavage band at 1 mM l-lysine is divided by the cleavage intensity in the absence of ligand (after background correction). (e) Plot depicting the normalized fraction of RNA cleaved (after correction for loading differences using the band of strong, lysine-independent cleavage annotated with ‘correction’) in region 1 for both labels (32P in black and Atto 633 in red) versus the concentration of l-lysine.