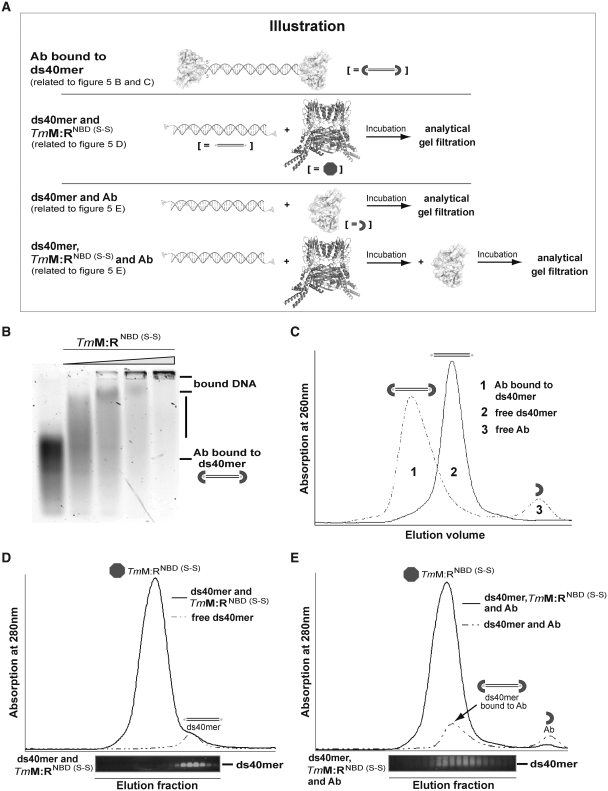
Figure 5.
Details of the DNA-binding mechanism of TmMRNBD (S-S) via the Antibody DNA-binding Assay. (A) Illustration of the Antibody DNA-binding Assay described in Figure 5B–E. (B) EMSA showing that TmMRNBD (S-S) in ATP/ADP state can still bind to antibody blocked dsDNA. Protein concentrations: 0, 1.75, 3.50, 7.0 and 14.0 µM. (C) Gel filtration chromatogram of 5′fluorescein labeled ds40mer in the absence (solid line) and presence (dashed line) of the antibody fragment FITC-E2 verified complete blocking of the respective dsDNA. The presence of antibody scFv shifted the DNA to larger molecular weights (peak 1) compared to DNA alone (peak 2) and free scFv (peak 3). (D) Gel filtration chromatogram of the 5′fluorescein labeled ds40mer in presence (solid line) and absence (dashed line) of TmMRNBD (S-S). Prior to gel filtration protein and DNA were incubated under conditions where in EMSA most of the DNA is shifted by bound protein. The gel filtration retention volumes were subsequently analyzed by agarose gel electrophoresis. The agarose gel lanes are aligned with the respective fractions of the gel filtration elution. (E) Gel filtration chromatograms of the DNA-antibody mixture (dashed line) and the ternary DNA-TmMRNBD (S-S)-antibody complex (solid line). Analysis of the respective elution fractions by agarose gel electrophoresis indicates that the complex could not be trapped on dsDNA.